![]() BS-SY-SC2-100BS-SY-SC2-1000
BS-SY-SC2-100BS-SY-SC2-1000
Bio-Speedy ® Direct RT-qPCR SARS-CoV-2Instructions For UseFor In vitro Diagnostic UseRx OnlyFor use under Emergency Use Authorization (EUA) only

![]() Bioeksen R&D Technologies Inc.Istanbul Technical University, Teknokent ARI 3, No: 4/B 105Sariyer, Istanbul, TURKEYPhone: +90 (212) 285 10 17, Fax: +90 (212) 285 10 18E-mail: [email protected].tr, Web: www.bioeksen.com.tr
Bioeksen R&D Technologies Inc.Istanbul Technical University, Teknokent ARI 3, No: 4/B 105Sariyer, Istanbul, TURKEYPhone: +90 (212) 285 10 17, Fax: +90 (212) 285 10 18E-mail: [email protected].tr, Web: www.bioeksen.com.tr
INTENDED USE
Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 nucleic acid detection kit is a one-step reverse transcription and real-time RT-PCR test intended for the qualitative detection of nucleic acid from SARS-CoV-2 in nasopharyngeal swabs, oropharyngeal (throat) swabs, combined nasopharyngeal/oropharyngeal swabs, anterior nasal swabs, mid-turbinate nasal swabs, nasal or nasopharyngeal aspirates, nasal washes and bronchoalveolar lavage samples from individuals suspected of COVID-19 by their healthcare provider. Testing is limited to laboratories certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a, that meet requirements to perform high complexity tests.Results are for the identification of SARS-CoV-2 RNA. The SARS-CoV-2 RNA is generally detectable in respiratory specimens during the acute phase of infection. Positive results are indicative of the presence of SARS-CoV-2 RNA; clinical correlation with patient history and other diagnostic information is necessary to determine patient infection status. Positive results do not rule out bacterial infection or co-infection with other viruses. The agent detected may not be the definite cause of the disease. Laboratories within the United States and its territories are required to report all results to the appropriate public health authorities.Negative results do not preclude SARS-CoV-2 infection and should not be used as the sole basis for patient management decisions. Negative results must be combined with clinical observations, patient history, and epidemiological information.The Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 is intended for use by qualified clinical laboratory personnel specifically instructed and trained in the techniques of real-time PCR and in vitro diagnostic procedures. The Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 is only for use under the Food and Drug Administration’s Emergency Use Authorization.
TEST PRINCIPLE
The Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 nucleic acid detection kit is a real-time reverse transcription-polymerase chain reaction (rRT -PCR) test. The SARS-CoV-2 primer and probe set(s) is designed to detect nucleic acid from the SARS-CoV-2 in nasopharyngeal swabs, oropharyngeal (throat) swabs, combined nasopharyngeal/oropharyngeal swabs, anterior nasal swabs, mid-turbinate nasal swabs, nasal or nasopharyngeal aspirates, nasal washes, and bronchoalveolar lavage samples from patients suspected of COVID-19 by their healthcare provider.Detection with the kit is achieved via rapid nucleic acid extraction from the respiratory tract samples followed by multiplex real-time RT-PCR targeting the SARS-CoV-2 specific ORF1ab gene and human RNase P gene and mRNA in real-time PCR instruments that are equipped with FAM and HEX detection channels.The oligonucleotide set targeting the human RNase P gene and mRNA functions as a control of the sampling, nucleic acid extraction, and inhibition. The kit also contains negative and positive control templates.
The kit contains vNAT ® buffer that extracts and preserves viral nucleic acids in respiratory tract samples. The event ® component enables the initiation of the real-time RT- PCR within 5 minutes of the introduction of the sample. Polyethyleneimine coated tetradecyl dimethyl benzyl ammonium chloride-based nanoparticles (NP) and tween-20 in vNAT ® lyse envelope and nucleocapsid of SARS-CoV-2 and release the genome. NP, guanidinium thiocyanate and NaN3 in view ® preserve the integrity of the released genomes. BSA in vNAT ® is used as a PCR facilitator to compensate for the negative effects of PCR inhibitors.
INSTRUMENTS AND SOFTWARE
The Bio-Speedy® Direct RT-qPCR SARS-CoV-2 test is to be used with the Roche LightCycler ® 96, Bio-Rad CFX96 Touch™, Qiagen Rotor-Gene ® 5 Plex Real-Time PCR Systems and the accompanying software (Table 1).
Table 1. Instruments and their software validated with the kit
| Manufacturer | Instrument | Instrument Catalog No | Software | Software Catalog No |
| Roche | LightCycler® 96 | 5815916001 | LightCycler® 96 Application and Instrument Software | Included in5815916001 |
| Bio-Rad | CFX96 TouchT” | 1855195 | CFX MaestroTM Software | 12004110 |
| Qiagen | Rotor-Gene® 5 Plex Platform | 9001570 | Q-Rex Software v1.0 | Included in9001570 |
REAGENTS AND MATERIALS PROVIDED
Reagent Names and ConcentrationsThe kit contents and their formulation are provided in Table 2.Table 2. Kit contents
| Component | Amount III | Content | Intended I. Use | Box 121 | |
| 100 Rx ns | 1000 Huts | ||||
| ()lip Mix | I x250 nl_ | 2x 1250 gl, | ORFI ab Primer-1; 500 nM | SARS-CoV-2 Detection(ORFI ab gene) | Box1/2 |
| ORFI ab Primer-2; 500 nM | |||||
| ORFI ab Probe; 150 nM (FAM) | |||||
| RNase P Primer-1; 500 nM | IntemalControl (IC) (RNase P gene and mRNA) | ||||
| RNase P Primer-2; 500 nM | |||||
| RNase P Probe; 150 nM (HEX) |
| 2X Prime Script Mix | lx500 L;I | 4×1250 gL | DNA polymerase, dNTP mix, reaction buffer, reverse ranscnp ta see ndri bo nuclease inhibitor | One Step Real-Tune RT-PCR TimeMix | 1/2 |
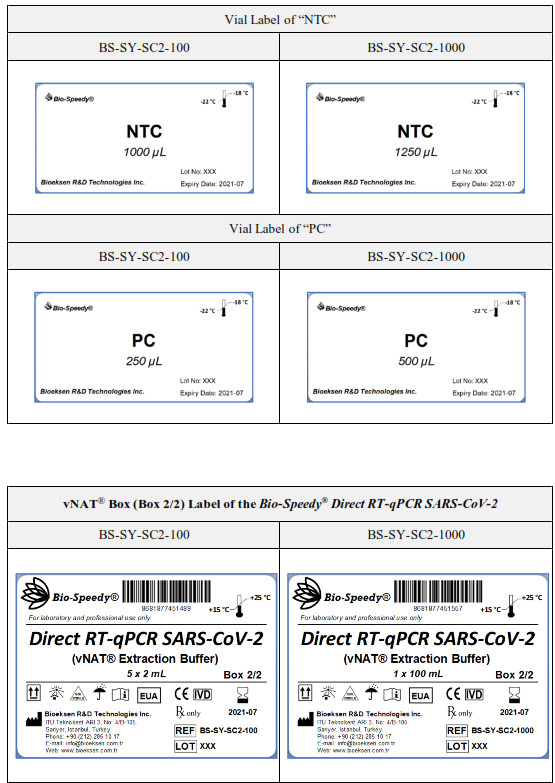
| NTC | 2×1000 gL | 16×1250 gL | Water, DEPC-Treated, MolecularBiology Grade (CAS 7732-18-5) | No Template (Negative)Control for PCR and Extraction Negative Control | Box in |
| PC | 1×250 gL | 2×500 gL | 103 copies/mL synthetic RNA for ORF I ab oligonucleotide set and 10 ng/gL total nucleic acid extract from human blood for RNase P oligonucleotide set | Positive Control Template: Synthetic SARSCoV-2 genome fragment | Box in |
| vNATe | 5×2 mL | lx100 mL | Viral nucleic acid extraction buffer | Extraction and preservation of viral nucleicacids | Box2/2 |
[1] 100-reactions kit Catalog No: BS-SY-SC2-100 and 1000-reactions kit Catalog No: BS-SY-SC2-1000.[2] Components with different storage and transport requirements are packaged in separate boxes. Please refer to the “LABELS” section.
Kit Storage Requirements and Shelf Life of the Kit
- Based on individual component shelf life, the approximate shelf life of the kit is estimated to be 12 months at the recommended storage temperature for each component.
- The “Box 1/2″ (PCR components: Oligo Mix, 2X Prime Script Mix, NTC and PC) can be stored at -20°C and can be transported at +2°C – +8°C. • The “Box 2/2” (Extraction component: vNAT ® ) can be stored and transported at room temperature (25°C ± 2°C). The temperature for long-term storage is +2°C- +8°C. However, it is recommended to be stored at room temperature because of the risk of precipitation occurring due to cold storage.
- Each reagent (stored at the recommended storage temperature) may be used until the expiration date indicated on the tube.
- The shelf life and safe storage conditions of the kit components were determined by the application of the Arrhenius Equation based on accelerated study data. During accelerated stability studies, all components were stored at different temperatures and tested at weekly intervals throughout the study period. More than 2 cycles of deterioration of Cq values for ORF1ab compared to the values obtained on day 0 was accepted as a threshold for the shelf life of the “2X Prime Script Mix”, “Oligo Mix” and “vNAT ® ” components. More than 5 cycles of deterioration of Cq values for ORF1ab compared to the values obtained on day 0 was accepted as a threshold for the shelf life of the “Positive Control (PC)” component.
- Real-Time stability testing is ongoing and the final shelf life will be established after the stability studies have been completed.
- The expiration date of the kit is determined by the expiration date of the included reagents and is indicated on the label of the kit box. In addition, the expiration date of each reagent is stated on the label of each reagent. Please refer to the expiration date on the box.
- Do not use reagents beyond their expiration date.
MATERIALS REQUIRED BUT NOT PROVIDED
Table 3. Components required but not included with the kit
[1] Any of these instruments may be used.
| Component | Using for | Specifications |
| Swabs for nasopharyngeal and oropharyngeal swab samples | Sampling and Transportation | Dacron or polyester, breakable shaft, sterile |
| Containers for other samples | Sampling and Transportation | Sterile, capped sample container |
| Viral Transport Medium | Sample transport | Preparation of VTM: Centers for Disease Control and Prevention, SOP#: DSR-052-01 |
| Vortex mixer | NA extraction | Speed up to 3000 rpm |
| Microcentrifuge tubes | NA extraction | 1.5 mL or 2 mL, nuclease-free |
| real-time RT-PCR | ||
| Micropipettes | NA extraction | Adjustable volume; 1-10 pL, 10-100 PI, 100-1000 PL |
| real-time RT-PCR | ||
| Micropipettes tips | NA extraction | Compatible with the micropipettes (I – 10 it, 10-100 pL and 100-1000 pL), filtered, nuclease-free |
| real-time RT-PCR | ||
| LightCyclers 96 with Application and Instrument Software III | real-time RT-PCR | Manufacturer: Roche / Catalogue No: 5815916001 |
| CFX96 Touch and CFXMaestroTM Software 1l1 | real-time RT-PCR | Manufacturer: Bio-Rad /Catalogue No: 1855195 (instrument) and 12004110 (software) |
| Rotor-Gene. 5 Plex Platform with Q-Rex Software v1.0111 | real-time RT-PCR | Manufacturer: Qiagen /Catalogue No: 9001570 |
| Reaction tubes and their caps/ seals | real-time RT-PCR | Compatible with the Real-Time PCR instrument and the reaction volume |
| Quick-spin centrifuge | real-time RT-PCR | Speed up to 3000 rpm, with adaptors for PCR plates and tubes |
| PCR cabinet | real-time RT-PCR | UV cabinet for PCR Setup |
| Cold tube rack | real-time RT-PCR | For microfuge tubes (1.5/ 2 mL) and for PCR tubes (0.1 / 0.2 mL) |
| Gloves | All processes | Disposable, powder-free, nitrite |
| Laboratory coat | All processes | Disposable (preferably) |
[1] Any of these instruments may be used.
WARNINGS AND PRECAUTIONS
Use Statements
- For In Vitro Diagnostic (IVD) Use only
- For prescription use (Rx) only
- For use under Emergency Use Authorization (EUA) only
- The Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 assay has not been FDA cleared or approved but has been authorized by FDA under an Emergency Use Authorization(EUA) for use in authorized laboratories – laboratories certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a, that meet requirements to perform high complexity tests.
- The Bio-Speedy® Direct RT-qPCR SARS-CoV-2 assay has been authorized only for the detection of nucleic acid from SARS-CoV-2, not for any other viruses or pathogens.
- The Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 assay is only authorized for the duration of the declaration that circumstances exist justifying the authorization of emergency use of in vitro diagnostic tests for detection and/or diagnosis of COVID-19 under Section 564(b)(1) of the Federal Food, Drug and Cosmetic Act, 21 U.S.C. § 360bbb-3(b)(1), unless the declaration is terminated or the authorization is revoked sooner.
- Laboratories within the United States and its territories are required to report all results to the appropriate public health authorities.
Waste Management
Medical WasteAppropriate waste management and decontamination procedures should be used.Medical wastes are collected in sealed biohazard bags/containers that are resistant to rupture, puncture, explosion, and transportation in accordance with the regulations on medical wastes.The contents of medical waste bags/containers should be never compressed, removed from the bag/container, emptied and transferred to another container.Dispose of waste in a designated matter in accordance with local, regional, and federal regulations.
Molecular Waste Nucleic acid contamination from molecular waste can be caused by dust and spreading aerosols.PCR products can be destroyed using a 3 % (mass fraction) hypochlorite solution (refer to ISO 22174:2005).
General Requirements for Good Practices on PCR and RT-PCR
Laboratory SetupTo prevent contamination of the reaction mixture by previously amplified target sequences, maintain separate work areas, dedicated equipment, and supplies for:
- Sample preparation
- PCR setup
- PCR amplification
- Analysis of PCR products
Rooms can be simulated using a clean bench or the UV cabinet for PCR setup. Physical separation through the use of different rooms is the most effective and preferable way of ensuring separate work areas and working facilities.For additional information, refer to ISO 22174 (2005).
Personnel
- Different sets of laboratory coats should be worn pre- and post-PCR.
- Disposable gloves should be worn at sample preparation and when setting up PCR.
- Laboratory coats and gloves should be changed at appropriate frequencies (when suspected that they are contaminated) and before leaving the work area (in passing from one work area to another).
- All personnel who perform aspects of the testing procedures should be trained to work with PCR and microbiology as appropriate.
Protection of Product Performance and Analysis Efficiency
- The components in the kit should not be mixed with components with different lot numbers or chemicals of the same name but from different manufacturers.
- Master stock reagents should be kept on the cold block during the PCR setup; if possible, the PCR setup should be performed on the cold block.
- Kit components should be mixed by gently shaking before use. Maintenance/ calibration intervals should be determined for all instruments and equipment used with the kit.
- Sampling should be carried out by personnel with sufficient knowledge and experience.
- For a collection of nasopharyngeal/ oropharyngeal swabs, polyester flocked swabs are preferred. Sterile dacron swabs with plastic or flexible metal handles may also be used. Cotton or calcium alginate swabs or swabs with wooden sticks should not be used since they may contain substances that inactivate some viruses and inhibit PCR.
- The kit is intended for use in a laboratory environment by qualified clinical laboratory personnel specifically instructed and trained in the techniques of real-time PCR and in vitro diagnostic procedures.
Preventing Contamination
- The kit should be stored away from nucleic acid sources and PCR amplicons.
- The micropipettes used for pipetting PCR mixes and template nucleic acids should be separate. Filtered and nuclease-free pipette tips should be used.
- All sample tubes should be opened and closed carefully to avoid contamination.
- Template nucleic acid and positive control tubes should always be kept closed, except for fluid transfers; tube caps should not be interchanged.
- Amplified products should not be brought into the reaction setup area. To avoid false positives due to amplified material, the PCR completed reaction tubes should be disposed of before opening in the laboratory (PCR products can be destroyed using a 3% -mass fraction- hypochlorite solution; refer to ISO 22174:2005).
- To avoid false positives due to cross-contamination, all unknown sample tubes should be closed before pipetting the Positive Control.
- It is recommended to use swabs with a breakable shaft to prevent contamination during sampling.
- The wipeable surfaces of the rooms, benches and devices where the analysis is performed should be cleaned regularly with freshly diluted 10% bleach solution (0.5% sodium hypochlorite).
Plate Layout Suggestions
- In multi-targeted PCR runs, separate different targets by a row or by a column if enough space is available.
- If possible, put at least one well between unknown samples and controls.
- Separate negative and positive controls by one well if possible.
- Place replicates of one sample for the same target next to each other.
- Start with the unknown samples and put controls at the end of the row or column.
- If possible, put positive controls in one of the outer rows or columns.
- Consider that caps for PCR tubes come in strips of 8 or 12.
SPECIMEN HANDLING AND STORAGE
Collecting the Specimen
Nasopharyngeal swabs, oropharyngeal (throat) swabs, combined nasopharynx ngeoro pharyngeal swabs, anterior nasal swabs, mid-turbinate nasal swabs, nasal or nasopharynx aspirates, nasal washes, and bronchial alveolar lavage samples shall be collected by a healthcare provider in accordance with the updated version of CDC Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens for COVID-19 (https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html).
Swabs (dacron or polyester flocked) should be placed immediately into a sterile transport tube containing 2-3 mL of viral transport medium (VTM) (Preparation of viral transport medium, Centers for Disease Control and Prevention, SOP#: DSR-052-01).Nasopharyngeal (NP) or nasal aspirate and nasal wash samples should be transferred into sterile containers containing 2-3 mL of VTM (in case of immediate analysis, these samples can be taken into sterile containers by healthcare providers).Bronchoalveolar lavage (BAL) samples should be collected 2-3 mL into a sterile, leak-proof, screw-cap sputum collection cup or sterile dry container.
Transporting Specimens
Specimens must be packaged, shipped, and transported according to the current edition of the International Air Transport Association (IATA) Dangerous Goods Regulation. Follow shipping regulations for UN 3373 Biological Substance, Category B when sending potential SARS-CoV2 specimens. Store specimens at 2-8°C and ship them overnight to the laboratory on an ice pack. If a the specimen is frozen at -70°C or lower, ship overnight to the laboratory on dry ice.
Storing Specimens
Specimens can be stored at 2-8ºC for up to 72 hours after collection. If a delay in extraction is expected, store specimens at -70ºC or lower in accordance with the CDC Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens for COVID-19. Extracted nucleic acid should be stored at -70ºC or lower. It is important to avoid repeated freezing and thawing of specimens.
INSTRUCTION FOR USE
Preparation of Nucleic Acid Samples
The following procedures are used to extract nucleic acid from nasopharyngeal swabs, oropharyngeal (throat) swabs, combined nasopharyngeal/ oropharyngeal swabs, anterior nasal swabs, mid-turbinate nasal swabs, nasopharyngeal or nasal aspirates, nasal washes in the VTM and from bronchoalveolar lavage samples.For the negative extraction control, the same procedures are applied using 100 µL of NTC in the kit instead of the respiratory sample.
- Vortex the sample tube at the highest speed for 15 seconds.
- Transfer 100 µL of vNAT ® into a clean microcentrifuge tube.
- Add 100 µL respiratory sample to the tube containing 100 µL vNAT®
- Vortex the tube at the highest speed for 15 seconds.
- Incubate the tube for 5 minutes at room temperature.
- The 200 µL mixture is ready to use in Real-Time RT-PCR.
- Store the sample at -70°C.
Planning of the PCR plate & PCR Setup
- Determine the number of reactions and create the PCR plate plan.
- Plan to include the following reactions:o Duplicate reactions for each test sample and extraction negative control.o Duplicate PCR control reactions;– Positive Control (included in the kit)– No Template (Negative) Control (included in the kit)
- Thaw all reagents.
- Vortex all reagents to mix thoroughly.
- Centrifuge the reagent tubes briefly to bring the contents to the bottom and place them on a cold rack/ice.
- Combine the following components for the number of reactions required plus 10% overage to compensate for pipetting errors:
Table 4. Reaction set-up
| Component | Volume per reaction[1] | Volume for 96 reactions [2] |
| 2X Prime Script Mix | 5 µL | 528 µL |
| Oligo Mix | 2.5 µL | 264 µL |
[1] These amounts are valid for a reaction volume of 10 µL. For 20 µL reaction volume, these values should be multiplied by 2.[2] Includes 10% overage to compensate for pipetting errors.
- Mix the solution thoroughly by vortexing, centrifuge briefly and distribute 7.5 µL (distribute 15 µL for the reaction volume of 20 µL) to each reaction well or tube.
- Add 2.5 µL (add 5 µL for the reaction volume of 20 µL) of Nucleic Acid sample, Extraction Negative Control, Negative (No Template) Control and Positive Control to the appropriate wells.
- Seal the plate or close the tubes, centrifuge briefly to bring the contents to the bottom, and place them into the Real-Time PCR instrument.
Programing and Running the Real-Time PCR Instrument
• The kit is validated for 10 µL and 20 µL Real-Time PCR volumes using Roche LightCycler ® 96, Bio-Rad CFX96 Touch™, and Qiagen Rotor-Gene ® 5 Plex Real-Time PCR Systems. It is recommended to use one of these instruments.• Program the Real-Time PCR instrument as follows:Table 5. Real-Time PCR program details
| Reaction Volume III | Step | CycleNumber | Temperature | Duration |
| 10 p L or 20µL | Reverse Transcription | 1 | 52 °C | 5 min |
| Hold | 1 | 95 °C | 10 sec | |
| Denature | 40 | 95 °C | 1 sec | |
| Anneal / Extend | 55 °C | 30 sec | ||
| Detection (Reading) | FAM (ORF lab) I HEX (RNase P) |
QUALITY CONTROL
- Quality control requirements must be performed in conformance with local, state, and federal regulations or accreditation requirements and the user’s laboratory’s standard quality control procedures. For further guidance on appropriate quality control practices, refer to 42 CFR 493.1256.
- Quality control procedures are intended to monitor reagent and assay performance.
- Test all positive controls prior to running diagnostic samples with each new kit lot to ensure all reagents and kit components are working properly.
- Good laboratory practice (GLP) recommends including a positive extraction control in each nucleic acid isolation batch.
- The negative extraction control must proceed through nucleic acid isolation per batch of specimens to be tested.
- Always include a negative control (NTC), and the appropriate positive control (PC) in each amplification and detection run. All clinical samples should be tested for the human RNase P (RP) gene to control for specimen quality and extraction.
Description of Controls
Controls that will be provided with the test kit are provided in Table 6.Table 6. Controls to be used with Bio-Speedy ® Direct RT-qPCR SARS-CoV-2
| Control 1) pe | Description | PurposeFrequency of | Testing |
| No Template (Negative) Control / Extraction Negative Control | Molecular grade, DNase and RNase-free water | Contamination control during RNA extraction and RT-PCR batch (100 µL) | Each extraction and each PCR run (2.5111, or 5 gla per reaction) |
| Positive Control | 103 copies/mL (=2-10x LoD) synthetic SARS- CoV-2 RNA and 10 ng/itL total nucleic acid extract from human blood which contains RNase P gene | To monitor the integrity of the RT-PCR reagents and process. | Each PCR run (2.5 id, or 5 FiL per reaction) |
| Internal/ Extraction Control | RNase P gene and d mRNA transcript (RNase P gene and mRNA targetedeted primers and probes areincluded in Oligo-Mix). | To monitor the integrity of specimen adequacy and the integrity of nucleic acid extraction and RTPCR from each human respiratory tract specimen | Every collected human respiratory tract specimen |
RESULT INTERPRETATION
Interpretation of Control Results
All test controls should be examined prior to the interpretation of patient results. If the controls are not valid, the patient results cannot be interpreted.No Template (Negative) Control/ Extraction Negative Control (NTC)The NTC is molecular grade, DNase and RNase-free water, used in place of sample nucleic acid as a no template (negative) control in each PCR run. NTC also serves as an extraction negative control and it is used in place of respiratory sample in each extraction batch/run. No Template (Negative) Control/ Extraction Negative Control should give a negative result (Cq not detected) for both the oligo mixes targeting ORF1ab (SARS-CoV-2, FAM channel) and RNase P (IC, HEX channel). Otherwise, it shows that there is a contamination problem. In this case, it is recommended to repeat the analysis by paying attention to the “Warnings and Precautions” section.
Positive Control
The Positive Control (PC) includes synthetic SARS-CoV-2 RNA that contains ORF1ab sequence and total nucleic acid extract from human blood which contains RNase P gene and mRNA. The positive control should give positive results (Cq<38.0) for both the oligo mixes targeting ORF1ab (SARS-CoV-2, FAM channel) and RNase P (IC, HEX channel). Otherwise, it indicates that there is a reagent stability problem. In this case, it is recommended to contact the manufacturer and renew the reagents and repeat the analysis.
Internal/ Extraction Control
Detection of RNase P in extracted nucleic acid serves as an extraction, inhibition and sampling control for each sample. Nucleic acid extracted from each specimen, should yield a positive result in the HEX channel, with a Cq value < 38.0. If the RNase P assay is negative on a clinical sample, it is interpreted as follows:• If the ORF1ab assay is positive with a negative RNase P result, it is considered that there is no inhibition, extraction or sampling problem and the run is valid. In this case,the result is interpreted as “SARS-CoV-2 Positive” as long as there is no sigmoidal amplification curve in the No Template (Negative) Control and the Positive Control isvalid.• If the ORF1ab assay is negative along with a negative RNase P, the specimen result is considered invalid and should be repeated. If the residual specimen is available, nucleic acidis re-extracted from the specimen, and the test is performed again. If the re-tested sample does not give a positive result in the HEX channel, a new specimen should be collected from the patient.Table 7. Expected Performance of Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 Controls
| Control Type | Control C Name | Purpose | Expected Results and Cq Values | |
| ORFlab | RNase P | |||
| No Template (Negative) Control / Extraction Negative Control | NTC | Contamination control during RNA extraction and RT-PCR | Negative(No Cq) | Negative(No Cq) |
| Positive Control | PC | Reagent integrity | Positive(Cq<38.0) | Positive(Cq<38.0) |
| Internal / Extraction Control | IC | To monitor the integrity of nucleic acid extraction and RT-PCR from each human respiratory tract specimen | Notapplicable | Positive(Cq<38.0) |
If any control does not perform as described above, run is considered invalid and all specimens are repeated from the extraction step. The positive control and negative control are interpreted as described in Table 8 below.
Table 8. Positive and Negative Control Interpretation
| Positive Control | Negative Control | Results | Action | ||
| ORFlab(FAM) | RNase P(HEX) | ORFIab(FAM) | RNase P(HEX) | ||
| Positive(Cq<38.0) | Positive(Cq<38.0) | Negative(No Cq) | Negative(No Cq) | VALID | Continue to result in the interpretation of patient specimens |
| Any of them is Negative(Cq not detected) | Not considered | INVALID(Reagent integrityproblem) | Contact the manufacturer, renew the reagents, and repeat the reaction | ||
| Not considered | Any of them isPositive(Cq<38.0) | INVALID(Contaminationproblem) | RRepeat the analysis by paying attention to the ” Warnings and Precautions” section. |
Examination and Interpretation of Patient Specimen Results
Assessment of clinical specimen test results must be performed after the positive and negative controls have been examined and determined to be valid and acceptable. If the controls are not valid, the patient results cannot be interpreted.The assay results are interpreted as given in Table 9. The results can be interpreted as “SARS-CoV-2 Positive” as long as there is no sigmoidal amplification curve in the negative control.The results can be interpreted as “SARS-CoV-2 Negative” as long as there is a sigmoidal amplification curve with a Cq<38.0 in the internal and positive controls.Table 9. Interpretation of Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 Patient Samples
| ORFIab / FAM (positive for Cq <38.0) | RNase P / HEX (positive for Cq <38.0) | Results Interpretation | Action |
| Positive (+) | Positive (+) | Results are VALID, SARS-CoV-2 RNA is Detected | Report as POSITIVE |
| Positive (+) | Negative (-) | Results are VALID, SARS-CoV-2 RNA is Detected | Report as POSITIVE |
| Negative (-) | Positive (+) | Results are VALID,SARS-CoV-2 RNA is Not Detected | Report as NEGATIVE |
| Negative (-) | Negative (-) | Results are INVALID(Sampling / Extraction / Inhibition problem) | Re-extract the specimen and perform testing again. If the result is still invalid, should be about new a specified. If additional the clinical sample is unavailable, report as INVALID |
PERFORMANCE CHARACTERISTICS
Limit of detection (LoD)
Limit of detection (LoD) studies determines the lowest detectable concentration of SARS-CoV2 at which greater or equal to 95% of all (true positive) replicates test positive.
In LoD studies, natural respiratory tract specimens that were RT-PCR negative for SARS-CoV2 collected from healthy donors and patients were used.
A cultured SARS-CoV-2 virus of an isolate from a patient (provided by the Republic of Turkey, Ministry of Health, General Directorate of Public Health) was used for spiking. The cultured virus was quantified in copies/mL by a molecular assay using dilutions of synthetic SARSCoV-2 ORF1ab gene partial RNA.
The samples for the LoD study were obtained by spiking natural respiratory samples, which were RT-PCR negative for SARS-CoV-2, with the cultured SARS-CoV-2 virus that was quantified previously. For each sample type, a total of 4 concentration levels with 1.5, 2 and 3fold dilutions between the levels were tested with a total of 120 replicates (20 replicates in 3 different instruments; Roche LightCycler ® 96, Bio-Rad CFX96 Touch™, Qiagen Rotor-Gene ® 5 Plex and at 2 different reaction volumes; 10 µL and 20 µL) per concentration, with an additional 60 replicates of blank samples (negative clinical specimens for SARS-CoV-2). The results of the LoD study for Nasopharyngeal Dacron Swab (NPDS) samples are summarized in Table 10. Similar results were obtained in the LoD study performed at both 10 µL and 20 µL reaction volumes using 3 different instruments.Table 10. Results of the LoD study for the Nasopharyngeal Dacron Swab (NPDS) samples
| Instrument | Reaction
Volume |
Concentr.of Viral RNA(copies/mL) | NumberTested | SARS-CoV-2 (ORF 1 ab) | Internal Control (RNase P) | |||
| Bio-Rad CFX96 Touch™ | 10 µL | Positive | al(Avg) | SD | Positive | Cq(Avg) | SD | |
| 200 | 20 | 20 (100.0%) | 37. | 0.03 | 20 (100.0%) | 34. | 0.08 | |
| 133 | 20 | 18 (90.0%) | 37. | 0.24 | 20 (100.0%) | 33. | 0.04 | |
| 100 | 20 | 11 (55.0%) | 38. | 0.04 | 20 (100.0%) | 34. | 0.04 | |
| 66 | 20 | 4 (20.0%) | 38. | 0.03 | 20 (100.0%) | 34. | 0.04 | |
| 0 | 10 | 0 (0.0%) | – | – | 10 (100.0%) | 34. | 0.03 | |
| 20 µL | 200 | 20 | 20 (100.0%) | 37. | 0.04 | 20 (100.0%) | 34. | 0.09 |
| 133 | 20 | 18 (90.0%) | 37. | 0.23 | 20 (100.0%) | 34. | 0.05 | |
| 100 | 20 | 10 (50.0%) | 38. | 0.03 | 20 (100.0%) | 34. | 0.03 | |
| 66 | 20 | 4 (20.0%) | 38. | 0.03 | 20 (100.0%) | 34. | 0.06 | |
| 0 | 10 | 0 (0.0%) | – | – | 10 (100.0%) | 34. | 0.05 |
| Instrument | ReactionVolume | Concentr.of Viral RNA(copies/mL) | Number | SARS-CoV-2 (ORF lab) | Interns Control (RNase P) | ||||
| Tested Positive | (Avg)(Avg) | SD | Positive | (Avg) | SD | ||||
| Roche LightCycler
® 96 |
10 µL | 200 | 20 | 20 (100.0%) | 37. | 0.04 | 20 (100.0%) | 34. | 0.15 |
| 133 | 20 | 17 (85.0%) | 37. | 0.23 | 20 (100.0%) | 34. | 0.11 | ||
| 100 | 20 | 12 (60.0%) | 38. | 0.04 | 20 (100.0%) | 34. | 0.02 | ||
| 66 | 20 | 5 (25.0%) | 38. | 0.03 | 20 (100.0%) | 34. | 0.03 | ||
| 0 | 10 | 0(0.0%) | – | – | 10(100.0%) | 34. | 0.08 | ||
| 20 µL | 200 | 20 | 20 (100.0%) | 37. | 0.04 | 20 (100.0%) | 34. | 0.07 | |
| 133 | 20 | 17 (85.0%) | 37. | 0.24 | 20 (100.0%) | 34. | 0.08 | ||
| 100 | 20 | 11 (55.0%) | 38. | 0.03 | 20 (100.0%) | 34. | 0.02 | ||
| 66 | 20 | 4 (20.0%) | 38. | 0.03 | 20 (100.0%) | 34. | 0.02 | ||
| 0 | 10 | 0 (0.0%) | – | – | 10 (100.0%) | 34. | 0.07 | ||
| Qiagen Rotor-Gene
® 5 Plex |
10 µL | 200 | 20 | 20 (100.0%) | 37. | 0.03 | 20 (100.0%) | 34. | 0.16 |
| 133 | 20 | 17 (85.0%) | 37.00 | 0.07 | 20 (100.0%) | 34. | 0.12 | ||
| 100 | 20 | 11 (55.0%) | 38. | 0.03 | 20 (100.0%) | 34. | 0.06 | ||
| 66 | 20 | 4 (20.0%) | 38. | 0.02 | 20 (100.0%) | 34. | 0.08 | ||
| 0 | 10 | 0 (0.0%) | – | – | 10 (100.0%) | 33.80 | 0.06 | ||
| 20 µL | 200 | 20 | 20 (100.0%) | 37. | 0.04 | 20 (100.0%) | 34. | 0.20 | |
| 133 | 20 | 17 (85.0%) | 37. | 0.24 | 20 (100.0%) | 34. | 0.07 | ||
| 100 | 20 | 11 (55.0%) | 38. | 0.03 | 20 (100.0%) | 34. | 0.05 | ||
| 66 | 20 | 4 (20.0%) | 38. | 0.02 | 20 (100.0%) | 34. | 0.03 | ||
| 0 | 10 | 0 (0.0%) | – | – | 10 (100.0%) | 34. | 0.08 |
LoD of the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 is 200 copies/mL for nasopharyngeal aspirate; 281 copies/mL for bronchoalveolar lavage; 562 copies/mL for oropharyngeal swab; 89 copies/mL for nasopharyngeal swab samples collected with polyester flocked swabs; 200 copies/mL for nasopharyngeal swab samples collected with dacron swabs. The summary of the LoD study stratified per specimen type is provided in Table 11.
Table 11. The summary of LoD study results for each specimen type
| Sample Type | Concentrationof Viral RNA(copies/mL) | Number | SARS-CoV-2(ORFIab) | Internal Control(RNase P) | ||||
| TestedPositive | (Avg)(Avg) | SD | Positive | (Avg) | SD | |||
| Nasopharyngeal Aspirate (NPA) | 200 | 120 | 120 (100.0%) | 36. | 0.08 | 120 (100.0%) | 34. | 0.04 |
| 133 | 120 | 107 (89.2%) | 37. | 0.18 | 120 (100.0%) | 34. | 0.07 | |
| 100 | 120 | 52 (43.3%) | 37. | 0.12 | 120 (100.0%) | 34. | 0.05 | |
| 66 | 120 | 25 (20.8%) | 38. | 0.09 | 120 (100.0%) | 35. | 0.05 | |
| 0 | 60 | 0 (0.0%) | – | – | 60 (100.0%) | 34. | 0.04 | |
| Bronchoalveolar Lavage (BAL) | 562 | 120 | 120 (100.0%) | 36. | 0.05 | 120 (100.0%) | 29. | 0.12 |
| 374 | 120 | 120 (100.0%) | 37. | 0.04 | 120 (100.0%) | 29. | 0.09 | |
| 281 | 120 | 116 (96.7%) | 37.30 | 0.05 | 120 (100.0%) | 29. | 0.10 | |
| 187 | 120 | 79 (65.8%) | 38. | 0.11 | 120 (100.0%) | 29. | 0.08 | |
| 0 | 60 | 0 (0.0%) | – | 60 (100.0%) | 29. | 0.04 | ||
| NasopharyngealDacron Swab
(NPDS) |
200 | 120 | 120 (100.0%) | 37. | 0.04 | 120 (100.0%) | 34. | 0.12 |
| 133 | 120 | 104 (86.7%) | 37. | 0.21 | 120 (100.0%) | 34. | 0.08 | |
| 100 | 120 | 66 (55.0%) | 38. | 0.03 | 120 (100.0%) | 34. | 0.04 | |
| 6O | 120 | 25 (20.8%) | 38. | 0.04 | 120 (100.0%) | 34. | 0.04 | |
| 0 | 60 | 0 (0.0%) | – | – | 60 (100.0%) | 34. | 0.06 | |
| Nasopharyngeal Polyester Flocked
Swab (NPFS) |
89 | 120 | 120 (100.0%) | 37. | 0.07 | 120 (100.0%) | 30. | 0.05 |
| 59 | 120 | 104 (86.7%) | 37. | 0.05 | 120 (100.0%) | 30. | 0.08 | |
| 44 | 120 | 57 (47.5%) | 38. | 0.11 | 120 (100.0%) | 30. | 0.09 | |
| 29 | 120 | 33 (27.5%) | 38. | 0.04 | 120 (100.0%) | 30.10 | 0.04 | |
| 0 | 60 | 0 (0.0%) | – | 60 (100.0%) | 30. | 0.07 | ||
| Oropharyngeal Dacron Swab (OPDS) | 562 | 120 | 120 (100.0%) | 37. | 0.04 | 120 (100.0%) | 35. | 0.06 |
| 374 | 120 | 110 (91.7%) | 37. | 0.03 | 120 (100.0%) | 35. | 0.02 | |
| 281 | 120 | 50 (41.7%) | 38. | 0.04 | 120 (100.0%) | 35. | 0.04 | |
| 187 | 120 | 28 (23.3%) | 38. | 0.03 | 120 (100.0%) | 35. | 0.04 | |
| 0 | 60 | 0 (0.0%) | – | – | 60 (100.0%) | 34.60 | 0.05 | |
| Oropharyngeal Polyester Flocked
Swab (OPFS) |
562 | 120 | 120 (100.0%) | 37. | 0.04 | 120 (100.0%) | 35. | 0.05 |
| 374 | 120 | 104 (86.7%) | 37. | 0.03 | 120 (100.0%) | 35. | 0.04 | |
| 281 | 120 | 55 (45.8%) | 38. | 0.03 | 120 (100.0%) | 35. | 0.06 | |
| 187 | 120 | 29 (24.2%) | 38. | 0.03 | 120 (100.0%) | 35. | 0.04 | |
| C) | 60 | 0 (0.0%) | – | 60 (100.0%) | 35. | 0.04 |
The effect of Real-Time PCR volume on test performance was also examined during LoD studies. Average Cq values of 10 µL and 20 µL Real-Time PCRs in LoD concentrations for each sample type were given in Table 12. These results showed that both 10 µL and 20 µL PCR volumes can be used for the assay.Table 12. The effect of Real-Time PCR volume on test performance
| SampleType | Viral RNA copA(1-1°D) | SARS-CoV-2 (ORF lab) | Internal Control (RNase P) | ||||||
| 20µL | 10µL | 20 A | 10µL | ||||||
| oi(Avg) | SD | Cq(Avg) | SD | Cq(Avg) | SD | Cq(Avg) | SD | ||
| NPA | 200 | 36. | 0.09 | 36. | 0.06 | 34. | 0.03 | 34. | 0.05 |
| BAL | 281 | 37. | 0.05 | 37. | 0.05 | 29. | 0.09 | 29. | 0.10 |
| NPDS | 200 | 37. | 0.04 | 37. | 0.03 | 34. | 0.11 | 34. | 0.14 |
| NPFS | 89 | 37. | 0.06 | 37. | 0.06 | 30. | 0.06 | 30. | 0.05 |
| OPDS | 562 | 37. | 0.04 | 37. | 0.04 | 34.80 | 0.06 | 35. | 0.06 |
| OPFS | 562 | 37. | 0.04 | 37. | 0.04 | 35. | 0.07 | 35. | 0.03 |
Inclusivity
All SARS-CoV-2 nucleotide sequences in available nucleotide databases(NCBI; https://blast.ncbi.nlm.nih.gov/Blast.cgi? ROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) were searched against the SARS-CoV-2 targeted oligonucleotide sequences. The results are summarized in Table 13. 99.7% and 99.9% of the available 15324 SARS-CoV-2 nucleotide sequences resulted in 100% identity for the forward and reverse primers respectively; 99.8% of the available 15324 SARS-CoV-2 nucleotide sequences resulted in 100% identity for the probe sequence. The maximum number of mismatches is 2 for the 25base long probe, 1 for the 22-base long forward primer, and 1 for the 28-base long reverse primer.
Table 13. Inclusivity est results (in-silico tests were carried out on August 27, 2020.)
| Oligo Name | MinimumIdentity111 | MaximumMismatches121 | Number of Total Alignments | Number of Alignments withMismatches | Number of Alignments with 100% Identity |
| ORFlab Primer-1 | 95.5% | 1 | 15324 | 41 | 15283 (99.7%) |
| ORFlab Primer-2 | 96.4% | 1 | 15324 | 13 | 15311 (99.9%) |
| ORFlab Probe | 92.0% | 2 | 15324 | 23 | 15301 (99.8%) |
[1] Minimum identity%= [(base long of the oligonucleotide – number of maximum mismatches) / base long of the oligonucleotide] x 100.[2] The decrease in Tm values when there are 1 base mismatch in the primer sequences or 2 base mismatches in the probe sequence do not prevent the primers and probe from remaining attached during the amplification step.
Exclusivity/ Cross-Reactivity
Wet Testing
The exclusivity was wet tested with a total of 44 samples, consisting of 43 respiratory pathogens at clinically relevant concentrations (10 5 genome copies/mL) and a pooled, SARS-CoV-2 nasal wash from 20 different people, following extraction by using vNAT ® . All samples were tested in duplicate and none produced any detectable reactivity with the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2.
The wet tests showed that the kit does not cross-react with the other respiratory pathogens or the microbial flora in the human respiratory tract (Table 14). In-Silico TestsIn silico tests were carried out using Primer Blast tool of NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) by entering the tested cross-reacting organism/strain and oligonucleotide sequences into the relevant fields. BLAST searchhttps://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome was also used for comparing the whole target region on SARS-CoV-2 Ref Seq Genome (NC_045512, 15431-15532) with the genomes of cross-reacting organisms/strains (Table 14 and Table 15). The SARS-CoV-2 taxon was excluded in the BLAST searches while the cross-reacting organisms/strains were included.The in-silico tests showed that the primers and probe were not homologous to any organism/strain except some SARS-CoV strains other than SARS-CoV-2 (Table 14). The blast search showed that the target region on the SARS-CoV-2 genome resembles more than 90% to some SARS-CoV strains, however, detection is not expected as there are no known SARS-CoV strains circulating in the population (Table 15).Table 14. The exclusivity (cross-reactivity) test results of 43 respiratory pathogens and a pooled nasal wash sample from 20 different people
| Organism | I n-SilicoAnalysis | Wet Testing (lir copies/mL) | |||
| ORFlab | RNase P | ||||
| Cq (Avg)111 | SD | Cq (Avg) III | SD | ||
| Human Coronavirus 229E | Not detected | No Cq | – | 31.49 | 0.02 |
| Human Coronavirus 0C43 | Not detected | No Cq | – | 31.51 | 0.05 |
| Human Coronavirus NL63 | Not detected | No Cq | – | 31.50 | 0.06 |
| Human Coronavirus HKUI | Not detected | No Cq | – | 31.46 | 0.03 |
| MERS-coronavirus | Not detected | No Cq | – | 31.51 | 0.01 |
| SARS CoV strain Frankfurt I | Not detected | No Cq | – | 31.45 | 0.04 |
| Influenza A H1 | Not detected | No Cq | – | 31.44 | 0.06 |
| Organism | In-Sliy ko Analsis | Wet Testing (105 copies/nil ) | |||
| ORFlab | RA’ase P | ||||
| Cq (Avg)III | SD | Cq (Avg)III | SD | ||
| Influenza A H3 | Not detected | No Cq | 32. | 0.10 | |
| Influenza B | Not detected | No Cq | 32. | 0.07 | |
| Parainfluenza I | Not detected | No Cq | 32. | 0.03 | |
| Parainfluenza 2 | Not detected | No Cq | 31. | 0.01 | |
| Parainfluenza 3 | Not detected | No Cq | 31. | 0.06 | |
| Parainfluenza 4 | Not detected | No Cq | 31. | 0.05 | |
| Human Metapneumovirus (hMPV) | Not detected | No Cq | 31.40 | 0.06 | |
| Rhinovirus | Not detected | No Cq | 31. | 0.01 | |
| Respiratory syncytial virus (RSV) A | Not detected | No Cq | 32. | 0.07 | |
| Respiratory syncytial virus (RSV) B | Not detected | No Cq | 31.50 | 0 | |
| Bocavirus (BoV) | Not detected | No Cq | 32. | 0.04 | |
| Enterovirus | Not detected | No Cq | 32. | 0.06 | |
| Adenovirus | Not detected | No Cq | 31. | 0.06 | |
| Legionella pneumophila | Not detected | No Cc! | 31. | 0.05 | |
| Chlamydia pneumonia | Not detected | No Cq | 31. | 0.09 | |
| AOrobactaium tuberculosis | Not detected | No Cq | 31. | 0.07 | |
| Haemophilus influenzae | Not detected | No Cq | – | 31. | 0.07 |
| Streptococcus pneumonia | Not detected | No Cq | – | 32. | 0.05 |
| Mycoplasma pneumonia | Not detected | No Cq | – | 31. | 0.04 |
| Streptococcus pyogenes | Not detected | No Cq | 31. | 0.04 | |
| Bordetella pertussis | Not detected | No Cq | 32. | 0.02 | |
| Pneumocystisjiroverii | Not detected | No Cq | 31. | 0.11 | |
| Candida Albicans | Not detected | No Cq | 31.50 | 0.04 | |
| Legionella bozemanii | Not detected | No Cq | 32. | 0.01 | |
| Legionella micdadei | Not detected | No Cq | 32. | 0.05 | |
| Corynebacterium diphtheriae | Not detected | No Cq | 31. | 0.05 | |
| Bacillus anthracis | Not detected | No Cq | 32. | 0.07 | |
| Moraxella catarrhalis | Not detected | No Cq | 31. | 0.06 | |
| Neisseria meningitidis | Not detected | No Cq | 32. | 0.08 | |
| Pseudomonas aeruginosa | Not detected | No Cq | 31. | 0.01 | |
| Staphylococcus epidennidi | Not detected | No Cq | 31. | 0.08 | |
| Coxiella but | Not detected | No Cq | 32. | 0.06 | |
| Staphylococcus aureus | Not detected | No Cq | 32. | 0.03 | |
| Streptococcus salubrious | Not detected | No Cq | 31. | 0.08 | |
| Leptospira interrogan | Not detected | No Cq | 31. | 0.04 |
| Organism | In-S111coAnalysis | NN’et Testing (105 copies/nil ) | |||
| ORFI ab | RA’ase P | ||||
| Cq (Avg)III | SD | Cq (Avg)III | SD | ||
| Chlamydia psittaci | Not detected | No Cq | – | 32. | 0.07 |
| Pooled human nasal wash – to represent diverse microbial floor in the human respiratory (met | – | No Cq | – | 28. | 0.08 |
[1] Average Cq values of duplicate PCRs in 10 5 copies/mL concentrations for each organism inVTM:Sample (1:1) mixtures.
Table 15. In-silico test results of the potential cross-reacting SARS-CoV strains
| Description | Accession | density % | ||
| Forward | Probe | Reverse | ||
| Rh Edolphus bat coronavirus BtCoV/4991 RNA-dependent RNA polymerase (RdRp) gene, partial CDs | KP876546. I | 100.% | 96.% | 100.% |
| Bat coronavirus RaTGI3, complete genome | MN996532. 1 | 100.% | 96.% | 100.% |
| Bat SARS-like coronavirus isolate 151569 RNA-dependent RNA polymerase (RdRp) gene, partial CDs | MN312711.I | 96.% | 100.0A | 92.% |
| Bat SARS-like coronavirus isolate 8548 RNA-dependent RNA polymerase (RdRp) gene, partial CDs | MN3 I 2738.1 | 96.% | 100.0A | 92.% |
| Bat SARS-like coronavirus isolate 8561 RNA-dependent RNA polymerase (RdRp) gene, partial CDs | MN3 I 2739.1 | 96.% | 100.0A | 92.% |
| Bat SARS-like coronavirus isolate 8572 RNA-dependent RNA polymerase (RdRp) gene, partial cds | MN312740.1 | 96.% | 100.0A | 92.% |
| Bat SARS-like coronavirus isolate 8586 RNA-dependent RNA polymerase (RdRp) gene, partial cds | MN3 I 2741.1 | 96.% | 100.0A | 92.% |
| Bat coronavirus Rc_CoV-3 RdRp gene for RNA dependent RNA polymerase, partial CDs | LC469044.1 | 100.% | 96.% | 92.% |
| SARS bat coronavirus RdRp gene for RNA dependent RNA polymerase, partial CDs. grain: 1s68 | A8889999.1 | 96.% | 96.% | 92.% |
| Bat SARS coronavirus Rp3. complete genome | DQ071615. I | 96.% | 96.% | 92.% |
| Bat SARS CoV Rs672/2006. complete genome | F3588686. I | 96.% | 96.% | 92.% |
| SARS-related bat coronavirus isolate Anlong-29 RNA-dependent RNA polymerase gene. partial CDs | KF294439. I | 96.% | 96.% | 92.% |
| SARS-related bat coronavirus isolate Anlong-97 RNA-dependent RNA polymerase gene. partial CDs | KF294440. I | 96.% | 96.% | 92.% |
| SARS-related bat coronavirus isolate Anlong-111 orflab polyprotein and orf I a polyprolein genes. complete CDs | ICF294455.1 | 96.% | 96.% | 92.% |
| Rh inolophus affinis coronavirus isolate LY Ra3 RNA- | ICF569973.1 | 96.% | 96.% | 92.% |
| Description | Accession | Identity % | ||
| Forward | Probe | Reverse | ||
| dependent RNA polymerase gene, partial cds | ||||
| Rhinolophus affinis coronavirus isolate Lyra I I, complete genome | KF569996. I | 96.% | 96.% | 92.% |
| Bts-BetaCoV/GX2013, complete genome | KJ473815. I | 96.% | 96.% | 92.% |
| SARS-related coronavirus isolate F21 RdRP mRNA, partial cds | KU973686.1 | 96.% | 96.% | 92.% |
| SARS-related betacoronavirus Rp3/2004 PREDICTEHA-156-12-HZI3479 RNA-dependent RNA polymerase mRNA, partial cds | 10:285180.1 | 96.% | 96.% | 92.% |
| SARS-related betacoronavirus Rp3/2004 PRE DICTE HA-156-12-HZ13484 RNA-dependent RNA polymerase mRNA, partial cds | 10:285181.1 | 96.% | 96.% | 92.% |
| Bat SARS-like coronavirus isolate Rf4092, complete genome | KY417145.1 | 96.% | 96.% | 92.% |
| Bat SARS-like coronavirus isolate Rs4231, complete genome | KY41 7146.1 | 96.% | 96.% | 92.% |
| Bat coronavirus isolate Anlong-103, complete genome | KY770858. I | 96.% | 96.% | 92.% |
| Bat coronavirus isolate Anlong-112, complete genome | KY770859.1 | 96.% | 96.% | 92.% |
| Bat coronavirus Rc_CoV-4 RdRp gene for RNA dependent RNA polymerase, partial cds | LC469045. I | 96.% | 96.% | 92.% |
| Coronavirus BtRs-BetaCoV/YN2018A, complete genome | MK211375.1 | 96.% | 96.% | 92.% |
| Bat SARS-like coronavirus isolate 8794 RNA-dependent RNA polymerase (RdRp) gene, partial CDs | MN3I2742.1 | 96.% | 96.% | 92.% |
| Bat SARS-like coronavirus isolate HZ13479 RNA-dependent RNA polymerase (RdRp) gene, partial cds | MN312829.1 | 96.% | 96.% | 92.% |
| Bat SARS-like coronavirus isolate HZ13484 RNA-dependent RNA polymerase (RdRp) gene, partial cds | MN312830.1 | 96.% | 96.% | 92.% |
| Bat SARS-like coronavirus isolate HZ13488 RNA-dependent RNA polymerase (RdRp) gene, partial cds | MN3I2831.1 | 96.% | 96.% | 92.% |
| Bat SARS-like coronavirus isolate NL140346 RNA-dependent RNA polymerase (RdRp) gene, partial cds | MN312843.1 | 95.5%92.0% | 96.% | |
| Bat SARS-like coronavirus isolate NL140352 RNA- dependent RNA polymerase (RdRp) gene, partial cds | 312844. | 96.% | 96.% | 92.% |
| Bat SARS-like coronavirus isolate NL140494 RNA- dependent RNA polymerase (RdRp) gene, partial cds | 312856. | 96.% | 96.% | 92.% |
Endogenous Interference Substances Studies
Nasopharyngeal dacron swab samples that were qPCR negative for SARS-CoV-2 collected from healthy donors and patients were used in the interference substances studies. Eight (8) NPDS samples, each in 2 mL of VTM, were pooled to obtain a total of 16 mL of negative clinical nasopharyngeal dacron swab sample. For each interfering substance listed in Table 16, 1 mL of negative NPDS sample was aliquoted into microcentrifuge tubes. 1 mL of sample in each tube was spiked with 50% of the relevant interfering substance. From each SARS-CoV-2 negative NPDS sample containing 50% interfering substance, 200 µL dilutions were prepared with final interfering substance concentrations of 10%, 1%, 0.1%, and 0.01%, as shown in Table 16. All dilutions were spiked with synthetic SARS-CoV-2 RNA to a final concentration of 1000 copies/mL. Nucleic acid was extracted from all samples using the vent ® buffer in BioSpeedy ® irect RT-PCR SARS-CoV-2 and then real-time RT-PCR was performed with the kit.According to the results of the interfering substance inhibition tests, mucin at 50% (w/v), blood at 50% (v/v), nasal spray (Nasonex) at 10% (v/v), nasal corticosteroids and gels t 10% (w/v), throat lozenges at 10% (w/v), anti-viral at 1% (v/v), antibiotics at 0.1% (w/v) may interfere with the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2.Table 16. Interfering substance inhibition of the Real-Time RT-PCR assay on the triplicate samples containing 1000 SARS-CoV-2 copies/mL
| Interfering substances | Concentrate ion | ORF I ab | !Was° P | Results(positive forCq < 38.0) | |||
| Cq111 | SD”‘ | Cqiii | SDP’ | ||||
| No interfering substances | 0 | 35. | 0.03 | 33. | 0.05 | Positive (+) | |
| Mucin from bovine submaxillary glands (CAS Number 84195-52-8) | w/v % | 0.I | 35. | 0.06 | 33. | 0.03 | Positive (+) |
| 1 | 35. | 0.04 | 33. | 0.02 | Positive (+) | ||
| 10 | 36. | 0.11 | 34. | 0.04 | Positive (+) | ||
| 50 | 39. | 0.08 | 37. | 0.13 | Negative(-) | ||
| Blood | v/v % | 0.I | 35. | 0.06 | 33. | 0.05 | Positive (+) |
| 1 | 35. | 0.03 | 33. | 0.08 | Positive (+) | ||
| 10 | 36. | 0.05 | 34. | 0.06 | Positive (+) | ||
| 50 | 39. | 0.05 | 37. | 0.07 | Negative (-) | ||
| OTRIVINE Adult Nasal Spray | v/v % | 0.1 | 35. | 0.08 | 33. | 0.07 | Positive (+) |
| I | 35. | 0.09 | 33. | 0.09 | Positive (+) | ||
| 10 | 36. | 0.05 | 34. | 0.09 | Positive (+) | ||
| 50 | 37. | 0.10 | 35. | 0.10 | Positive (+) | ||
| Nasonex Nasal Spray | v/v % | 0.1 | 35. | 0.02 | 33. | 0.10 | Positive (+) |
| 1 | 36. | 0.06 | 34. | 0.05 | Positive (+) | ||
| 10 | 40. | 0.06 | 38. | 0.09 | Negative (-) | ||
| 50 | N/Am | – | N/A131 | – | Negative (-) |
| Interfering substances | Concentration | ORFIab | RNase P | Results(positive forCq < 38.0) | |||
| aim | spin | Cqu I | SDP’ | ||||
| Ayr Saline Nasal Gel, WithSoothing Aloe | w/v % | 0.1 | 35. | 0.08 | 33. | 0.06 | Positive (+) |
| 1 | 35. | 0.06 | 33. | 0.06 | Positive (+) | ||
| 10 | 39. | 0.07 | 37. | 0.08 | Negative(-) | ||
| 50 | N/Alm | – | N/A’3′ | – | Negative (-) | ||
| Strepsils Sore Throat &Blocked Nose Lozenges | 1 0/w v ° | 0.01 | 35. | 0.04 | 33. | 0.04 | Positive (+) |
| 0.1 | 35. | 0.02 | 33. | 0.08 | Positive (+) | ||
| 1 | 37. | 0.04 | 35. | 0.07 | Positive (+) | ||
| 10 | 40. | 0.07 | 38. | 0.14 | Negative(-) | ||
| Rapivab (peramivir) | v/v % | 0.01 | 35. | 0.03 | 33. | 0.09 | Positive (+) |
| 0.1 | 36. | 0.03 | 34. | 0.06 | Positive (+) | ||
| 1 | 39. | 0.07 | 37. | 0.12 | Negative (-) | ||
| 10 | N/API | – | N/A’31 | – | Negative(-) | ||
| Amoxicillin + Penicillin + Cefadroxil + Erythromycin mixture | w/v % | 0.01 | 36. | 0.12 | 34. | 0.05 | Positive (+) |
| 0.1 | 40. | 0.09 | 38. | 0.09 | Negative(-) | ||
| 1 | N/API | – | Nam | – | Negative(-) | ||
| 10 | N/An’ | – | N/A’3′ | – | Negative (-) |
[1] The average Cq value for triplicate reactions.[2] “SD” is the standard deviation of results for triplicate samples.[3] N/A: Not available; not detected any Cq value for all triplicate reactions.
FDA SARS-CoV-2 Reference Panel Testing
The evaluation of sensitivity and MERS-CoV cross-reactivity was performed using reference material (T1), blinded samples, and a standard protocol provided by the FDA. The study included a range-finding study and a confirmatory study for LoD. Blinded sample testing was used to establish specificity and to corroborate the LoD. RNA extraction from the samples was performed using the event ® buffer included in the kit (the extraction method with the event ® buffer is described in this IFU under the title Preparation of Nucleic Acid Samples”). The tests were carried out using the Bio-Rad CFX96 Touch™ Real-Time PCR instrument. The results are summarized in Table 17.Table 17. Summary of LoD Confirmation Result Using the FDA SARS-CoV-2 Reference Panel
| Reference MaterialsProvided by FDA | Specimen Type | Product LoD(NDU/mL) | Cross-Reactivity |
| SARS-CoV-2 | Nasopharyngeal DacronSwab | 1.8×103 | N/A |
| MERS-CoV | N/A | ND |
NDU/mL = RNA NAAT detectable units/mLN/A: Not ApplicableND: Not Detected
Clinical Evaluation
Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 was used for testing 451 clinical samples (nasopharyngeal swabs, oropharyngeal swabs, nasopharyngeal aspirates, and bronchoalveolar lavage samples) in VTM concurrently with another Real-Time RT-PCR kit authorized by the FDA in a blinded fashion. Samples were obtained from individuals suspected of COVID-19 (47%) or from individuals having COVID-19 (53%). No restrictions were placed on age, gender, medications or known pharmaceutical therapies. 451 individuals in the intensive care unit (ICU) (46%) and non-ICU settings (54%) were enrolled in this study.
Nasopharyngeal (NP) or oropharyngeal (OP) swab samples, NP aspirate, and bronchoalveolar lavage (BAL) samples were collected by a healthcare provider in accordance with the updated version of CDC Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens for COVID-19(https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html). Swabs (dacron or polyester flocked) were placed immediately into sterile transport tubes containing 2-3 mL of the viral transport medium (VTM). Nasopharyngeal (NP) aspirate and nasal wash samples were transferred into sterile containers containing 2-3 ml of VTM. Bronchoalveolar lavage (BAL) samples were collected 2-3 mL into sterile, leak-proof, screw-cap sterile dry containers. The samples were packaged and transferred to the “Turkish Ministry of Health, General Directorate of Public Health, Department of Microbiology Reference Laboratories and Biological Products, National Virology Reference Laboratory” in VTM at 2-8 °C within 24 hours for the SARS-CoV-2 real-time RT-PCR tests.As the samples arrived in the laboratory, they were processed immediately without being stored. The homogenized samples were divided into two separate tubes equally to be tested by two different methods.Nucleic acid extraction was carried out using vNAT ® buffer for the samples to be tested with Bio-Speedy ® Direct RT-qPCR SARS-CoV-2. The reactions were set up as described in Table 4 for the reaction volume of 10 µL. Real-Time RT-PCR tests were performed on Bio-Rad CFX96 Touch™ according to the PCR protocol as described in Table 5. Extracted nucleic acids were stored at -80ºC. To observe the effect of reaction volume in clinical samples, 451 samples were retested at 20 µL reaction volume, in accordance with Table 4 and Table 5, using frozen nucleic acid samples. Similar results were obtained at both 10 µL and 20 µL reaction volumes.The samples to be tested with the comparator method were treated as specified in the IFU of the relevant kit.The overall Bio-Speedy ® tests resulted in 349 true positives, 94 true negatives and 8 false negatives. The sensitivity and specificity of the Bio-Speedy ® Direct RT-qPCR SARS- CoV-2 are 97.8% and 100% respectively.
Table 18. Performance of the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 vs FDA authorized test
| Patient Specimens (for al sample types) | FDA authorized test | |||
| Positive | Negative | Total | ||
| Bio-Speedy® Direct RTqPCR SARS-CoV-2 | Positive | 349 | 0 | 349 |
| Negative | 8 PI | 94 | 102 | |
| Total | 357 | 94 | 451 | |
| Positive Percent Agreement | 349/357 = 97.8% (95% CI: 92.6% – 100%) | |||
| Negative Percent Agreement | 94/94 =100% (95% CI: 86.2% – 98.1%) |
[1] Positive by the comparator for the ORF1ab target only:
4 of the 8 samples were negative by the Bio-Speedy ® and positive by the comparator were negative by the DNA sequencing. The sequencing-based screening was performed by amplifying a 251 bp fragment of Orf 1ab using the following primer set: Cor-FW (5′-ACTCAAATGAATCTTAAGTATGC-3′) and Cor-RV (5′- TCACATTTTGGATAATCCCA-3′). The samples that were negative by DNA sequencing were a BAL sample and three NP / OP swab samples, the Cq values of which were 27.07, 28.56, 26.86 and 32.94, respectively, by comparative testing.
4 of the 8 samples were negative by the Bio-Speedy ® and positive by the comparator were positive by the DNA sequencing. Four of the sequenced 251 bp amplicons were 100% similar to the reference SARSCoV-2 (NC_045512) genome position between 15058 and 15308. The samples positive by the DNA sequencing were all NP / OP swab samples whose Cq values were determined as 23.55, 24.79, 28.96 and 30.61 by the comparative test.
ASSAY LIMITATIONS
- Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 is intended for use in a laboratory environment by qualified clinical laboratory personnel specifically instructed and trained in the techniques of real-time PCR and in vitro diagnostic procedures.
- The clinical specimens shall be collected by a healthcare provider in accordance with the updated version of CDC Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens for COVID-19 (https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelinesclinical-specimens.html).
- A false-negative result may occur if a specimen is improperly collected, transported or handled.
- Performance of the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 has only been established in the nasopharyngeal swab, oropharyngeal (throat) swab, nasopharyngeal aspirate, and bronchoalveolar lavage samples. Combined nasopharyngeal/oropharyngeal swabs, anterior nasal swabs, mid-turbinate nasal swabs, nasal aspirates and nasal washes are also considered acceptable specimen types but performance has not been established.
- The use of cotton or calcium alginate swabs or swabs with wooden sticks can lead to false-negative results since they may contain substances that inactivate some viruses and inhibit PCR. Flocked (polyester) or dacron swabs are recommended for the collection of nasopharyngeal/ oropharyngeal swab samples. The performance of the Bio-Speedy ® Direct RT-PCR SARS-CoV-2 has only been evaluated using Dacron and polyester flocked swabs.
- Mutations within the target regions of the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 could affect primer and/or probe binding resulting in failure to detect the presence of the virus.
- Inhibitors or other types of interference may produce a false negative result. Mucin at 50% (w/v), blood at 50% (v/v), nasal spray (Nasonex) at 10% (v/v), nasal corticosteroids and gels at 10% (w/v), throat lozenges at 10% (w/v), anti-viral at 1% (v/v), antibiotics at 0.1% (w/v) may interfere with the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2. False-negative results may also occur if inadequate numbers of organisms are present in the specimen.
- Detection of SARS-CoV-2 RNA may be affected by patient factors (e.g., presence of symptoms), and/or stage of infection.
- Based on the in-silico analysis, other SARS-like coronaviruses in the same subgenus (Sarbecovirus) as SARS-CoV-2 may cross-react with the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2. Other SARS-like coronaviruses in the same subgenus (Sarbecovirus) as SARS-CoV-2 are not known to be currently circulating in the human population, therefore are highly unlikely to be present in patient specimens.
- The clinical performance has not been established in all circulating variants but is anticipated to be reflective of the prevalent variants in circulation at the time and location of the clinical evaluation. Performance at the time of testing may vary depending on the variants circulating, including newly emerging strains of SARS-CoV-2 and their prevalence, which changes over time.
The Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 Letter of Authorization, along with the authorized Fact Sheet for Healthcare Providers, the authorized Fact Sheet for Patients, and authorized labeling are available on the FDA website:
However, to assist clinical laboratories using the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2, the relevant Conditions of Authorization are listed below:A. Authorized laboratories 1 using the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 must include with test result reports, all authorized Fact Sheets. Under exigent circumstances, other appropriate methods for disseminating these Fact Sheets may be used, which may include mass media.B. Authorized laboratories using the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 must use the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 as outlined in the authorized labeling. Deviations from the authorized procedures, including the authorized instruments, authorized extraction methods, authorized clinical specimen types, authorized control materials authorized other ancillary reagents and authorized materials required to use the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 are not permitted.C. Authorized laboratories that receive the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 must notify the relevant public health authorities of their intent to run the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 prior to initiating testing.D. Authorized laboratories using the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 must-have process in place for reporting test results to healthcare providers and relevant public health authorities, as appropriate.E. Authorized laboratories must collect information on the performance of the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 and report to Division of Microbiology (DMD)/Office of Health Technology 7 (OHT7)-Office of In Vitro Diagnostics and Radiological Health (OIR)/Office of Product Evaluation and Quality (OPEQ)/Center for Devices and Radiological Health (CDRH) (via email: [email protected]) and Bioeksen R&D Technologies Inc. (via e-mail: i[email protected] or via phone: +90 212 285 10 17) any suspected occurrence of false positive or false negative results and significant deviations from the established performance characteristics of the BioSpeedy ® Direct RT-qPCR SARS-CoV-2 of which they become aware.F. All laboratory personnel using the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 must be appropriately trained in RT-PCR techniques and use appropriate laboratory and personal protective equipment when handling this kit and use the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 in accordance with the authorized labeling.G. Bioeksen R&D Technologies Inc., its authorized distributor(s), and authorized laboratories using the Bio-Speedy ® Direct RT-qPCR SARS-CoV-2 must ensure that any records associated with this EUA are maintained until otherwise notified by FDA. Such records will be made available to FDA for inspection upon request.
1 The letter of authorization refers to, “Laboratories certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a, that meet requirements to perform high complexity tests” as “authorized laboratories.”
EXPLANATION OF SYMBOLS
Table 19. Symbols used in labeling for Bio-Speedy Direct RT-qPCR SARS-CoV-2

LABELS


MANUFACTURER and TECHNICAL SUPPORT
Bioeksen R&D Technologies Incorporated Company
| Address: | Istanbul Technical University Teknokent ARI 3, No: 4/B 105 Sariyer, Istanbul, TURKEY |
| Phone: | +90 (212) 285 10 17 |
| Fax: | +90 (212) 285 10 18 |
| Web: | www.bioeksen.com.tr |
| E-mail: | [email protected] |
BS-SY-SC2, IFU (Version May 24, 2021)
References
[xyz-ips snippet=”download-snippet”]