
SARS-CoV-2 Test


For use under the Emergency Use Authorization (EUA) onlyFor in vitro diagnostic useFor use with Nasal and Nasal Mid-Turbinate SwabsFor use with the Accula™ Dock and Silaris™ Dock
Instructions for Use
INTENDED USE
The Accula™ SARS-CoV-2 Test performed on the Accula Dock or the Silaris™ Dock is a molecular in vitro diagnostic test utilizing polymerase chain reaction (PCR) and lateral flow technologies for the qualitative, visual detection of nucleic acid from SARS-CoV-2 in clinician-collected nasal or nasal mid-turbinate swab specimens or clinician-instructed self-collected (collected on-site) nasal swab specimens, collected from individuals suspected of COVID-19 by their healthcare provider. Testing is limited to laboratories certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a, that meet requirements to perform high, moderate, or waived complexity tests. The Accola SARS-CoV-2 Test is authorized for use at the Point of Care (POC), i.e., inpatient care settings operating under a CLIA Certificate of Waiver, Certificate of Compliance, or Certificate of Accreditation.
Actual SARS-CoV-2 Test results are for the identification of SARS-CoV-2 RNA. The SARS-CoV-2 RNA is generally detectable in upperrespiratory specimens during the acute phase of infection. Positive results are indicative of the presence of SARS-CoV-2 RNA; clinical correlation with patient history and other diagnostic information is necessary to determine patient infection status. Positive results do notrule out bacterial infection or co-infection with other viruses. Testing facilities within the United States and its territories are required to report all results to the appropriate public health authorities.Negative results do not preclude SARS-CoV-2 infection and should not be used as the sole basis for patient management decisions.Negative results must be combined with clinical observations, patient history, and epidemiological information.The Accola SARS-CoV-2 Test is intended for use by trained operators who are proficient in performing tests on the Accula Dock andSolaris Dock. The Accola SARS-CoV-2 Test is only for use under the Food and Drug Administration’s Emergency Use Authorization. SUMMARY AND
EXPLANATION
The Accola SARS-CoV-2 Test performed on the Accula Dock or the Solaris Dock is a molecular in vitro diagnostic test utilizing polymerase chain reaction (PCR) and lateral flow technologies for the qualitative, visual detection of the coronavirus SARS-CoV-2 viral RNA. The actual SARS-CoV-2 Test uses a nasal or nasal mid-turbinate swab specimen collected from patients who meet CDC SARS-CoV-2 clinical criteria and in conjunction with epidemiological criteria to aid in the diagnosis of SARS-CoV-2 infection.
PRINCIPLE OF THE TEST
The Accola SARS-CoV-2 Test is a Nucleic Acid Amplification Test (NAAT) for detection of SARS-CoV-2 viral RNA in approximately 30minutes. To perform the test, nasal or nasal mid-turbinate specimens are added to the SARS-CoV-2 Buffer to solubilize the sample. An aliquot of the SARS-CoV-2 Buffer is then dispensed into an Accula SARS-CoV-2 Test Cassette. The Test Cassette contains internal process positive and negative controls, enzymes, OscAR™ reagents, and a detection strip necessary for the full completion of the assay.There are 4 steps in the process:
- Lysis of the virus
- Reverse transcription (RT) of viral RNA to cDNA
- Nucleic acid amplification by thermocycling polymerase chain reaction (PCR)
- Detection
The Accola Dock controls reaction temperatures, timing, and fluid movements within the Test Cassette resulting in a fast and automatedSARS-CoV-2 RT-PCR assay. After approximately 30 minutes, the test results are interpreted by the visualization of Blue Test Lines on the detection strip in the Test Cassette. A blue process control line at the control (C) area is used to ensure proper reagent and Accola Dock function and to confirm a valid negative test result.
REAGENTS AND MATERIALS MATERIALS PROVIDED
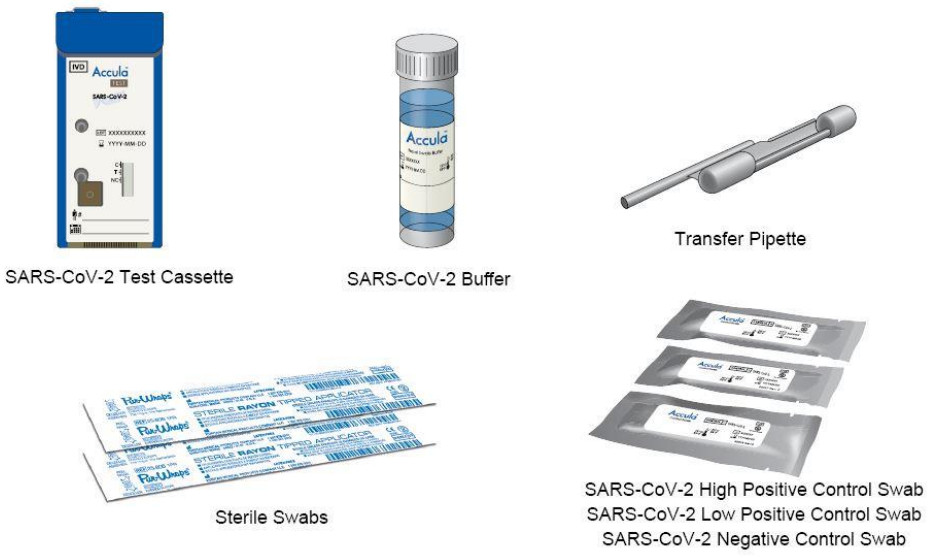
ACCULA SARS-CoV-2 TEST MATERIALS PROVIDED:
- Collection Swabs (25): Sterile swabs for nasal sample collection.
- SARS-CoV-2 Buffer (25): Single-use vial of a solution containing 5 mL of buffer with dimethyl sulfoxide and < 0.01% sodium azide.
- Transfer Pipette (25): Single-use, fixed volume pipette used to transfer the sample from the SARS-CoV-2 Buffer vial into the Test Cassette. NOTE: Supplied within the Test Cassette Pouch. Extra pipettes are provided for your convenience.
- Actual SARS-CoV-2 Test Cassette (25): Single-use, foil-pouched with desiccant and Test Cassette containing lyophilized reagents for the targeted amplification and detection of viral nucleic acid.
- SARS-CoV-2 High Positive Control Swab (1): DNA Based Synthetic Oligo dried onto a swab well above the limit of detection of the test.
- SARS-CoV-2 Low Positive Control Swab (1): DNA Based Synthetic Oligo dried onto a swab near the limit of detection of the test.
- SARS-CoV-2 Negative Control swab (1): buffer solution dried onto a swab.
- Self-Collection Quick Reference Guide (1)
- Electronic Instructions for Use (EU) Card (1)
- Instructions for Use and Quick Reference Guide is provided on the Mesa Biotech website, https://www.mesabiotech.com/aboutus/#product-documentation
MATERIALS PROVIDED SEPARATELY
- Actual Dock (Catalog # D2000) or Solaris Dock (Catalog #1026)
- Actual SARS-CoV-2 Control Kit (Catalog #COV4100-1)
STORAGE AND HANDLING
- Store reagents at room temperature (15°C to 30°C, 59°F to 86°F). Do not refrigerate or freeze.
- Do not reuse kit contents: Collection Swabs, Test Cassettes, Transfer Pipettes, Control Swabs, or SARS-CoV-2 Buffer.
- Do not remove the Test Cassette from the foil pouch until immediately before use (within 30 minutes).
- Do not use kits or reagents past the expiration date.
- Specimen swabs must be eluted in Accula SARS-CoV-2 Buffer immediately after sample collection.
- Eluted samples in Accula buffer may be kept at room temperature (15°C to 30°C, 59°F to 86°F) for up to 2 hours or refrigerated at 2°C to 8°C and tested within 24 hours from the time of elution.
- Eluted samples in Accula buffer may be stored for up to 1 week at -20°C; longer storage should be at -80°C or colder.
PRECAUTIONS
- For in vitro diagnostic use under Emergency Use Authorization only.
- This test has not been FDA cleared or approved but has been authorized for emergency use by the FDA for use by laboratories certified under the Clinical Laboratory Improvement Amendments (CLIA) of 1988, 42 U.S.C. §263a, that meet requirements to perform high, moderate or waived complexity tests. The test is authorized for use at the Point of Care (POC), i.e., inpatient care settings operating under a CLIA Certificate of Waiver, Certificate of Compliance, or Certificate of Accreditation.
- This test has been authorized only for the detection of nucleic acid from SARS-CoV-2, not for any other viruses or pathogens.
- The emergency use of this test is only authorized for the duration of the declaration that circumstances exist justifying the authorization of emergency use of in vitro diagnostics for the detection and/or diagnosis of COVID-19 under Section 564(b)(1) of the Federal Food, Drug, and Cosmetic Act, 21 U.S.C. § 360bbb-3(b)(1), unless the declaration is terminated or authorization is revoked sooner.
- Positive results are indicative of the presence of SARS-CoV-2 RNA.
- Laboratories within the United States and its territories are required to report all results to the appropriate public health authorities.
- To be used in conjunction with the Accola Dock or Solaris Dock.
- Follow universal precautions when handling patient samples. All patient samples should be treated as if potentially infectious.Follow standard BSL-2 guidelines when working with patient samples. Put on the appropriate personal protective equipment.
- If the buffer solution contacts the skin, wash the area with soap and clean water and rinse thoroughly. Consult a physician if irritation develops.
- DNA Based Synthetic Oligo is used to make the positive control swabs. However, Control Swabs, patient samples and Test Cassettes should be handled as though they could transmit disease. Observe established precautions against microbial hazards during use and disposal.
- Dispose of kit reagents and patient samples according to all local, state, and federal regulations.
- Do not use swabs or SARS-CoV-2 Buffer other than those provided with the Accula SARS-CoV-2 Test Kit.
- Do not write on the Test Cassette except in the indicated area on the Test Cassette label for recording sample identification and test date.
- Do not remove the foil tab from the Test Cassette until immediately before use. Once the tab is removed, add the sample immediately (within 5 minutes) and start testing.
- Once the sample is added and the Dock lid is closed, the test has started. Do not move the Dock, open the lid, or unplug the Dock until the Dock indicates the test has been completed.
- Do not use any damaged kit contents.
- Do not use kit components after their expiration date.
- Sample collection and handling procedures require specific training and guidance.
- All test kit components are single-use items. Do not use with multiple specimens.
- To help obtain accurate results, follow all instructions, and regard all precautions in this Instructions for Use.
- Inadequate or inappropriate sample collection, handling, processing, and/or storage can yield inaccurate results.
- Use only the fixed volume Transfer Pipette provided in the kit to transfer the patient sample from the Accula SARS-CoV-2 Buffer tube into the Test Cassette port. Do not pour the patient sample from the Accula SARS-CoV-2 Buffer vial into the Test Cassette sample port.
- Do not use visually bloody or overly viscous samples.
- When transferring the eluted patient sample, avoid drawing up large particulates, which may clog the Transfer Pipette.
- Due to the high sensitivity of the Accula SARS-CoV-2 Test, contamination of the work area with previous samples may cause false-positive results. Clean the Accula Dock or Solaris Dock and surrounding surfaces as described in the procedure in the Actual Dock or Solaris Dock Operators Guide.
- Do not attempt to open a used Test Cassette or a Test Cassette with a closed sample port.
- Do not touch the heads of the Control Swabs. Cross-contamination may occur due to the high sensitivity of the test.
- Use the Results Interpretation table in this Instructions for Use to interpret results accurately.
QUALITY CONTROL
Process ControlsEach Accola SARS-CoV-2 Test Cassette contains two internal process controls: an internal positive control (labeled ‘C’ on the Test Cassette) and negative control (labeled ‘NC’ on the Test Cassette). The positive process control is a non-infectious RNA bacteriophage in the Test Cassette and is used as the positive process control to verify assay steps (RNA extraction, reverse transcription, amplification, and detection) were executed properly. A non-SARS-CoV-2 nucleic acid probe is used as a negative control for false-positive results due to nonspecific binding.
Refer to the Interpretation of Results section of this Instruction for Use for instructions on interpreting the results for the Process Control.
External Positive and Negative ControlsExternal controls may be used to show that the Accula SARS-CoV-2 Test is working properly. The Accola SARS-CoV-2 Test kitcontains three Control Swabs:
- 1 SARS-CoV-2 High Positive Control Swab
- 1 SARS-CoV-2 Low Positive Control Swab
- 1 SARS-CoV-2 Negative Control Swab
- Mesa Biotech recommends that a SARS-CoV-2 negative and SARS-CoV-2 positive controls be run:
- Once for each new lot or shipment of kits received.
- Once for each new operator.
- As deemed additionally necessary to conform with your internal quality control procedures, with local, state, and/or federal regulations, or accrediting groups.
Additional Control Swabs may be purchased from Mesa Biotech (Catalog # COV4100-1). Run Control Swabs using the same procedure as for a patient specimen.If External QC testing fails, repeat the test using a new Control Swab, reagent, and Test Cassette, or contact Mesa Biotech Technical Support for assistance before testing patient samples.
SPECIMEN COLLECTION
Each test should be completed with a nasal swab sample using one SARS-CoV-2 Buffer vial.Proper sample collection is an important step for an accurate test result. Carefully follow the instructions below.NOTE: Use only the Collection Swabs supplied with the kit or one of the approved swab types listed below:
Nasal Swabs:
- Puritan Sterile Rayon Swab w/ Polystyrene Handle (REF. 25-806-1PR)
- Puritan Sterile Standard Foam Swab w/Polystyrene Handle (REF. 25-1506 1PF)
- Copan FLOQSwabs™o (REF. 502CS01) – Regular Size Nylon® Flocked Swab with 80mm Breakpointo (REF. 519CS01) – Regular Size Nylon® Flocked Swab with 100mm Breakpointo (REF. 520CS01) – Regular Size Nylon® Flocked Swab with 30mm Breakpoint
- McKesson Cotton Tip Wood Shaft 6 Inch Sterile (REF. 24-106-2S or REF. 24-106-1S)
- Dynarex Cotton Tipped Applicators (REF. 4305)
Nasal Mid-turbinate Swabs:
- Copan FLOQSwabs™o (REF. 56380CS01) – Contoured Adult Size Nylon® Flocked Swab with Stopper with 80mm Breakpointo (REF. 56780CS01) – Contoured Pediatric Size Nylon® Flocked Swab with Stopper with 80mm Breakpointo (REF. 56750CS01) – Contoured Pediatric Size Nylon® Flocked Swab with Stopper with 50mm Breakpoint
Nasal Swab SampleTo collect a nasal swab sample, insert a new sterile swab into the patient’s nostril until you feel slight resistance. Firmly rotate the swabagainst the nasal wall 10 times.Using the same swab, repeat this sampling procedure in the other nostril.Patient self-collection: On-site, supervised patient self-collection may be employed to reduce the risk of SARS-CoV-2 transmission between patients and testing site personnel. Patients may conduct nasal swab self-collection of themselves or a child, as described in the self-collection Quick Reference Guide. Testing site personnel must provide a physical or digital copy of the Self-Collection Quick Reference Guide to the patient prior to collection.Self-collection must occur on-site, under supervision by testing site personnel.Self-collection is limited to patients 18 years and older.Collection by an adult on a child should only be performed on children 5 years and older.Nasal swabs from children ages 0-4 years should be collected by the clinician.
NOTE: If the buffer solution contacts the skin, wash the area with soap and clean water and rinse thoroughly.Consult a physician if irritation develops.Nasal Mid-Turbinate Swab Sample To collect a nasal mid-turbinate swab sample, first, tilt the patient’s head back 70degrees.While gently rotating the mid-turbinate swab, insert the swab into the nostril until resistance is met at turbinates. Rotate the swab severaltimes against the nasal wall.
Using the same swab, repeat this sampling procedure in the other nostril.
- Sample Elution Specimen swabs must be eluted in Accula buffer immediately after sample collection.
- Patient nasal swabs previously stored in media other than Accula SARS-CoV-2 Buffer are not recommended and may yield invalid results or false results. For details, refer to the Limitations section.
| Write the patient identification (ID) information and testing date onto the SARS-CoV-2Buffer vial label in the area provided. |  |
| Insert the nasal swab specimen into the SARS-CoV-2 Buffer and rotate it 5 timesrubbing it against the wall of the vial.Remove the patient nasal swab from the SARS-CoV-2 Buffer vial and discard it into abiohazardous waste container.NOTE: If the buffer solution contacts the skin, wash the area with soap and clean water and rinse thoroughly. Consult a physician if irritation develops. | 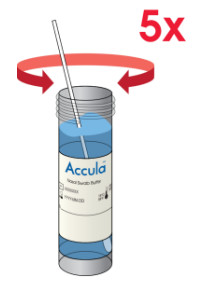 |
  |
TEST PROCEDURE
All clinical samples must be at room temperature before beginning the assay.Check the expiration date on the outer box before using it. Do not use any test after the expiration date on the box.
| Place Dock on a flat surface.Connect the AC Adapter to the Power Cord.Insert the round end of the power cord into the Dock. Plug the AC power cord into an electrical outlet. |   |
| Open the Dock by depressing the black button located on the top left. | 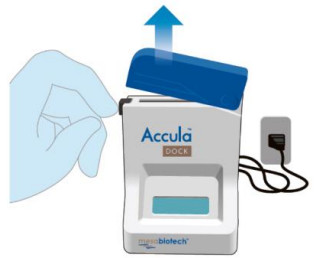  |
| Verify the Dock screen displays:“DOCK READY INSERT CASSETTE”. |   |
  |
|
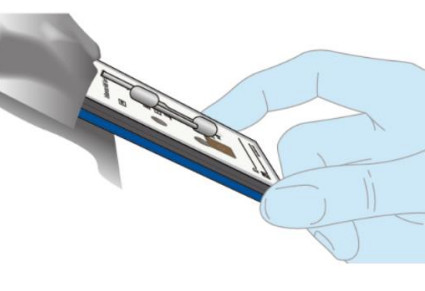
| Insert the Test Cassette into the Dock, leaving the lid open. Press the Test Cassette down firmly to seat it in the Dock.NOTE: Do NOT remove the foil tab covering the sample port until immediately before testing. |
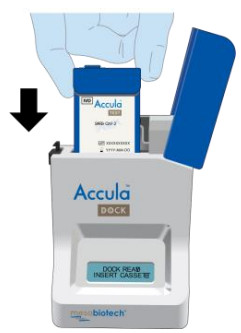  |
| Do not close the Dock lid until the sample has been added to the Test Cassette.Verify the Dock screen displays: “SARS-COV-2 CASS. INSERTED”The Dock screen will then display: “ADD SAMPLE THEN CLOSE LID” |   |
| Invert SARS-CoV-2 Buffer vial to mix.Remove the cap from the eluted patient sample in the SARS-CoV-2 Buff |   |
| Firmly squeeze the TOP bulb of the pipette.While continuing to squeeze the top bulb firmly, place the pipette tip will be the surface of the liquid in the SARS-CoV-2 Buffer vial.Keep the pipette tip well below the surface of the liquid of the vial containing the eluted patient sample in the SARS-CoV-2 Buffer vial.Slowly release the top bulb to completely fill the pipette stem with the sample.Some liquid may also be in the overflow reservoir.
NOTE: Although excess liquid will enter the pipette’s overflow chamber, only the liquid in the pipette stem will be dispensed. |
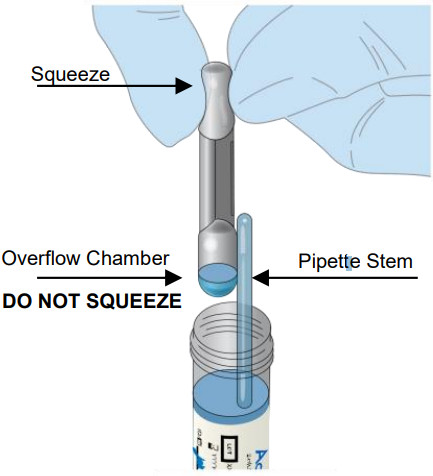  |
| Completely remove the foil tab covering the sample port on the Test Cassette. Discard the foil tab.NOTE: Once the tab is removed from the sample port, the sample must be added immediately (within 5 minutes). |   |
| Insert the tip of the pipette all the way into the sample port of the Test Cassette until resistance is met.Squeeze the TOP bulb of the pipette firmly to dispense all of the samples from the pipette stem into the Test Cassette.NOTE: A small amount of sample may remain in the overflow chamber (lower bulb). This is normal. | 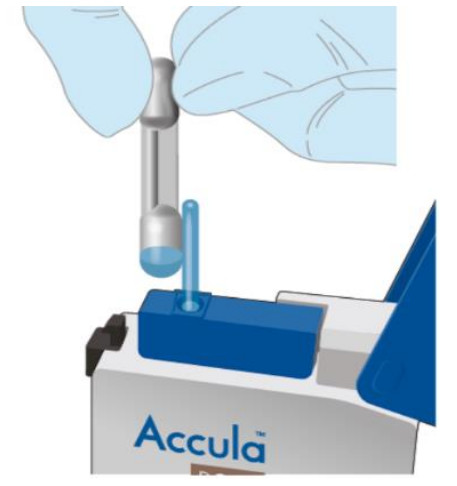  |
| Dispose of the pipette in a biohazardous waste container. | 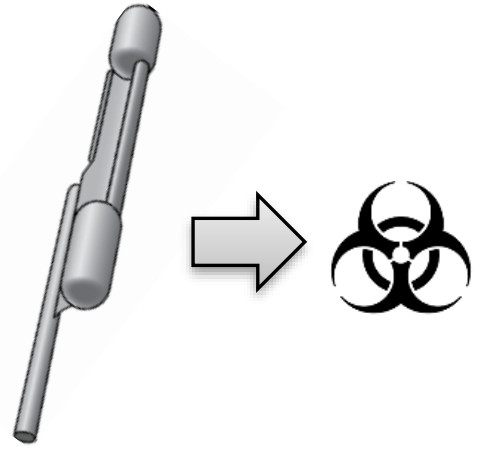  |
| The Dock screen will then read: “SAMPLE LOADED CLOSE LID”.Close the lid of the Dock immediately to automatically begin the test program. Verify the Dock screen displays: “SAMPLE LOADED LID CLOSED”. |   |
| Verify the Dock screen displays: “CASSETTE SEALED TEST STARTED”. |   |
| Verify the Dock screen displays: “TEST RUNNING REMAINING XX: XX”.Note: The test takes approximately 30 minutes to complete. The screen will continue to display “TEST RUNNING” until complete. The Dock will beep at the end of test processing. |
  |
| Verify the Dock screen displays: “TEST COMPLETE READ RESULTS”. |   |
| Open the lid of the Dock.Remove the Test Cassette and interpret the results according to the Interpretation of Results section below.Note: Results should be interpreted within 1 hour of test completion. | 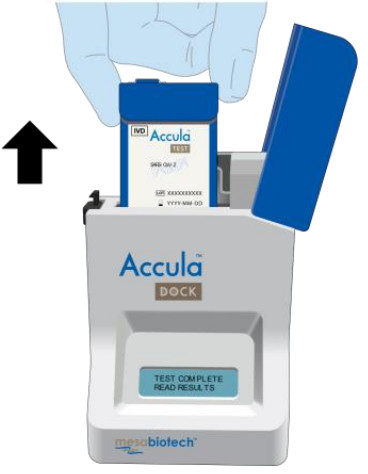  |
| Dispose of the Test Cassette in the biohazardous waste container. |   |
INTERPRETATION OF RESULT
NOTE: LOOK CLOSELY WHEN INTERPRETING RESULTS! The appearance of any shade of the Blue Test Line at the “T” position is a result that is interpreted as positive for SARS-CoV-2. A negative result will only contain a Blue Test Line at the “C” position.
C = Internal Positive Process ControlT = SARS-CoV-2NC = Internal Negative Process Control
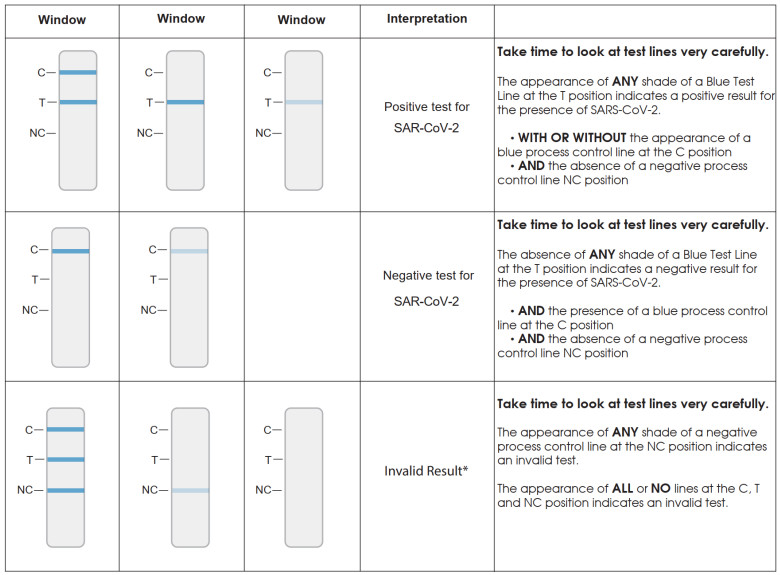

*If an invalid result is obtained, the sample may be rerun with a fresh Test Cassette only if the eluted sample in Accula buffer has been stored for less than 2 hours at room temperature. (15°C – 30°C or 59°F -86°F). Alternatively, a new sample should be collected and run with a new Buffer and Test Cassette.
NOTE: The absence of a Blue Test Line at the “C” position in conjunction with a Blue Test Line at the “T” position means that the SARS-CoV-2 target was amplified and detected as a valid result. This can occur due to the overabundance of the SARS-CoV-2 target thatcompetes with the Control target.
DOCK CLEANINGMesa Biotech recommends cleaning the Dock each day it is used.
Procedure:Clean the Accula or Solaris Dock and surrounding area according to the instructions provided in the cleaning section of the Accula Dock or Solaris Dock Operator’s Guide.
LIMITATIONS
- The performance of the Accula SARS-CoV-2 Test was determined using the procedures provided in this Instructions For Use.Failure to follow these procedures may alter test performance.
- The Accula™ SARS-CoV-2 Test is for use with nasal or nasal mid-turbinate swab specimens.
- Improper collection, storage, or transport of specimens may lead to false-negative or invalid results.
- Collection of patient samples into media other than the supplied Accula SARS-CoV-2 Buffer (such as UTM, VTM, or saline), or dilution of previously collected samples out of UTM, VTM, or saline into Accula SARS-CoV-2 Buffer is an off-label use and has beenshown to adversely impact test performance (“Comparison of the Accula SARS-CoV-2 Test with a Laboratory-Developed Assay for Detection of SARS-CoV-2 RNA in Clinical Nasopharyngeal Specimens,” Hogan, C.A., et al., J. Clin Microbiol. 2020 Aug; 58(8):e01072-20.)
- Test results should be interpreted in conjunction with the patient’s medical history, clinical signs, and symptoms, and the results of other diagnostic tests were performed.
- As with other tests, negative results do not rule out SARS-CoV-2 infections and should not be used as the sole basis for patient management decisions.
- Detection of SARS-CoV-2 RNA may be affected by sample collection methods, patient factors (e.g., presence of symptoms), and/or stage of infection.
- False-negative or invalid results may occur due to interference or the presence of inhibitors. The Internal Control is included, Accola SARS-CoV-2 Test to help identify the specimens containing interfering substances or inhibitors.
- This is a qualitative test. Test line intensity is not indicative of the quantity of virus in the sample.
- False-negative results may occur if viruses are present at levels below the test’s limit of detection.
- False-negative results may occur if mutations are present in the regions targeted by the test.
- Analytical studies evaluating the SARS-CoV-2 N gene 28881 GGG->AAC mutation have shown a potentially reduced sensitivity mayoccur when viral concentrations are at or near the limit of detection of the assay.
- Cross-reactivity with respiratory tract organisms other than those listed in the Analytical Specificity Study may lead to erroneous results.
- This test cannot rule out diseases caused by other viral or bacterial agents.
- Analyte targets (viral nucleic acid) may persist in vivo, independent of virus viability. Detection of analyte targets does not imply that the corresponding viruses are infectious, or are the causative agents for clinical symptoms.
Conditions of Authorization for LaboratoriesThe Accola SARS-CoV-2 Test Letter of Authorization, along with the authorized Fact Sheet for Healthcare Providers, the authorized FactSheet for Patients and authorized labeling are available on the FDA website: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/Vitro-diagnostics-as
To assist clinical laboratories using the Accula SARS-CoV-2 Test, the relevant Conditions of Authorization are listed below and are required to be met by laboratories and/or patient care settings performing the test.A. Authorized laboratories¹ using the Accula SARS-CoV-2 test must include with test result reports all authorized Fact Sheets. Under exigent circumstances, other appropriate methods for disseminating these Fact Sheets may be used, which may include mass media.B. Authorized laboratories using the Accula SARS-CoV-2 Test must perform the Accula SARS-CoV-2 Test as outlined in the Accola SARS-CoV-2 Test Instructions for Use. Deviations from the authorized procedures, including authorized instruments, authorized clinical specimen types, authorized control materials, authorized other ancillary reagents, and authorized materials required to perform the Accula SARS-CoV-2 Test, are not permitted.C. Authorized laboratories that receive the Accula SARS-CoV-2 Test must notify the relevant public health authorities of their intent to run the test prior to initiating testing.D. Authorized laboratories using the Accula SARS-CoV-2 Test must have a process in place for reporting test results to healthcare providers and relevant public health authorities, as appropriate.E. Authorized laboratories must collect information on the performance of the test and report to DMD/OHT7-OIR/OPEQ/CDRH (viaemail: [email protected]) and Mesa Biotech, Inc. Technical Support (via email: [email protected]) anysuspected occurrence of false positive or false negative results and significant deviations from the established performancecharacteristics of the test of which they have become aware.F. All operators using your product must be appropriately trained in performing and interpreting the results of your product, use appropriate personal protective equipment when handling this kit, and use your product in accordance with the authorized labeling.G. Mesa Biotech, its authorized distributors and authorized laboratories using the Accula SARS-CoV-2 Test must ensure that any records associated with this EUA are maintained until otherwise notified by FDA. Such records must be made available to FDA for inspection upon request.¹ The Letter of Authorization refers to “laboratories certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a, to high, moderate or waived complexity tests. This test is authorized for use at the Point of Care (POC), i.e., inpatient care settings operating under a CLIA Certificate of Waiver, Certificate of Compliance, or Certificate of Accreditation.” as “authorized laboratories”.
PERFORMANCE CHARACTERISTICS
Limit of Detection (LoD)
LoD testing was performed with inactivated SARS-CoV-2 virus from BEI (NR-52350). The preliminary LoD was determined by testing 5replicates with 2 fold dilutions. To confirm the LoD, dilutions were performed in pooled negative nasal swab human clinical matrix to identify the concentration that produced at least 95% positive results. Confirmatory testing was performed using three lots of Accola SARS-CoV-2 Test cassettes. Twenty (20) replicates from each cassette lot were tested with inactivated virus diluted in the clinical matrix.The LoD was confirmed to be 150 copies per mL.
| Virus | LoD (Copies per mL) | Positive/Replicates (%) |
| SARS-CoV-2 Inactivated Virus (BEI) | 150 | 58/60 (97%) |
Analytical Reactivity/InclusivityDue to the limited availability of SARS-CoV-2 isolates for inclusivity testing, in silico analysis was employed to evaluate the extent of homology between Accula SAR-CoV-2 primers and probes and all sequenced SARS-CoV-2 isolates from the United States available in the GISAID database as of December 5, 2020. 35,556 sequences were examined to identify the extent of predicted assay inclusivity. The table below summarizes the homology between 35,556 SARS-CoV-2 sequences and the Accula SARS-CoV-2 Test primers and probes.The forward primer shares 100% homology with 29612 of the 35556 available sequences (83.3% of sequences with perfect match). 4552SARS-CoV-2 sequences in the database (12.8% of sequences evaluated) carry the same 3 mismatches in the 5’ region of the forward primer (GGG->AAC) resulting in forwarding primer homology of 90.3% in these 4552 isolates. 99.5% of database sequences share a perfect match with the 6 bases of the forward primer 3’ terminus. The reverse primer binding sequence is consistently well conserved at all positions with greater than 97.8% of the database sequences sharing perfect homology with the reverse primer. 99.7% of databases sequences share a perfect match with the 6 bases of the reverse primer 3’ terminus.Amplicon detection in the Accula test cassette is accomplished through hybridization of amplicon to detect oligonucleotide conjugated to dyed polystyrene microspheres and to capture oligonucleotide probes immobilized on the detection strip at discrete line positions to generate a visible colorimetric signal. The detection probe is 100% homologous to 35435 of the 35556 database entries (99.7%) while the capture probe is similarly well-conserved sharing perfect homology with 35497 of the 35556 of the database entries (99.8%).
Table 1. In Silico Analysis of Inclusivity for the Accula SARS-CoV-2 Test
| Oligonucleotide | Homology Description |
| N gene Forward Primer | 29612 of 35556 complete SARS-CoV-2 genome sequences share 100% homology to the Forward Primer.35368 of 35556 complete SARS-CoV-2 genome sequences share 100% homologywith the 3’ terminal 6 bases of the Forward Primer.4552 of 35556 complete SARS-CoV-2 genome sequences carry 3 mismatches (GGG>AAC) in the 5’ portion of the Forward Primer resulting in 90.3% homology.Laboratory testing with sequences carrying this mutation show showed a potential for reduced sensitivity. Please see Table 2. |
| N gene Reverse Prime | 34768 of 35556 complete SARS-CoV-2 genome sequences share 100% homology to the Reverse Primer.35470 of 35556 complete SARS-CoV-2 genome sequences share 100% homologywith the 3’ terminal 6 bases of the Reverse Primer. |
| Detection Probe | 35435 of 35556 complete SARS-CoV-2 genome sequences share 100% homology to the Detection Probe. |
| Capture Probe | 35497 of 35556 complete SARS-CoV-2 genome sequences share 100% homology to the Capture Probe. |
In silico analysis revealed that the forward primer is predicted to be bound to the mismatched template at the annealing/extension temperatures of the assay.Table 2. Three Accola SARS-CoV-2 test cassette lots were used to test performance with a perfect primer match synthetic template (Perfect Match (GGG)) or a synthetic template carrying the variant sequence (Mismatch Template (AAC)).
| Results Summary for Three Cassette Lots at LoD | |||
| Concentration (copy/mL) | 1200 | ||
| Cassette Lot | 2011A099 | 2011C105 | 2011A1102 |
| Perfect Match Template (GGG) | 19/20 (95%) | 20/20 (100%) | 20/20 (100%) |
| Mismatch Template (AAC) | 20/20 (100%) | 17/20 (85%) | 18/20 (90%)* |
| “One invalid result was obtained and repeated, yielding a valid result |
Analytical Specificity – Exclusivity (Cross-Reactivity)The table below summarizes the findings of in silico exclusivity analysis examining the homology between the indicated organisms and the Accula SARS-CoV-2 Test primers and probes. Potential interactions are noted where primer homology exceeds 75%. SARS-CoV is the only organism identified as potentially cross-reactive by in silico analysis. While the primer binding sites are well conserved between sequenced isolates of SARS-CoV and SARS-CoV-2, the capture and detection probe binding regions share only 70% and 65% homologyrespectively. Probe homology with the consensus of a 226 sequence SARS-CoV alignment is listed in Table 3. Based only on sequence analysis, we cannot rule out the possibility that the Accula SARS-CoV-2 Test may cross-react with SARS-CoV. However, the low prevalence of SARS-CoV renders the observation of cross-reactivity unlikely. Indeed, SARS-CoV has not been detected in the human population since 2004.
In addition to in silico analysis, the Accula SARS-CoV-2 Test will be challenged with nucleic acids isolated from human coronaviruses OC43, NL63, HKU1 and 229E to confirm the test does not cross-react with these human coronaviruses.
In Silico Analysis of Exclusivity for the Accula SARS-CoV-2 Test
| Organism | Homology |
| SARS-CoV | Forward primer 93.5% homology with SARS-CoV ConsensusReverse primer 90.3% homology SARS-CoV ConsensusDetection probe 65% homology with SARS-CoV ConsensusCapture probe 70% homology with SARS-CoV Consensus |
| MERS-CoV | No alignment found |
| Human coronavirus 229E | No alignment found |
| Human coronavirus OC43 | No alignment found |
| Human coronavirus HKU1 | No alignment found |
| Human coronavirus NL63 | No alignment found |
| Adenovirus | No alignment found |
| Human Metapneumovirus | No alignment found |
| Parainfluenza virus 1-4 | No alignment found |
| Influenza A & B | No alignment found |
| Enterovirus | No alignment found |
| Respiratory Syncytial virus | No alignment found |
| Rhinovirus | No alignment found |
| Chlamydia pneumoniae | No alignment found |
| Haemophilus influenza | No alignment found |
| Legionella pneumonphila | No alignment found |
| Mycobacterium tuberculosis | No alignment found |
| Streptococcus pneumonia | No alignment found |
| Streptococcus pyogenes | No alignment found |
| Bordetella pertussis | No alignment found |
| Mycoplasma pneumonia | No alignment found |
| Pneumocystis jirovecii | No alignment found |
| Candida albicans | No alignment found |
| Pseudomonas aeruginosa | No alignment found |
| Staphylococcus epidermis | No alignment found |
| Staphylococcus salaries | No alignment found |
Exclusivity (Cross-Reactivity) Testing
The exclusivity study was performed by testing 32 potentially cross-reacting organisms with the Accula SARS-CoV-2 Test. Each organismwas diluted in a pooled negative human throat swab and nasal swab matrix and tested in triplicate. The organisms and concentrations are shown in the table below. None of the 32 organisms cross-reacted in the Accula SARS-CoV-2 Test at the concentrations tested.
Cross-Reactivity Testing for the Accula SARS-CoV-2 Test
| Target Organisms | Organism ReferenceNumber or strainavailable | Unit | ConcentrationTested | % Agreement with Expected Result |
| Adenovirus (e.g. C1 Ad. 71) | Type 1 | TCID50/mL | 5.10E+06 | 100% (3/3) |
| Human Metapneumovirus (hMPV) | IA14-2003 | TCID50/mL | 1.00E+05 | 100% (3/3) |
| Parainfluenza Type 1 | Type1 | TCID50/mL | 1.00E+05 | 100% (3/3) |
| Parainfluenza Type 2 | Type2 | TCID50/mL | 1.00E+05 | 100% (3/3) |
| Parainfluenza Type 3 | Type3 | TCID50/mL | 1.00E+05 | 100% (3/3) |
| Influenza A | Texas | TCID50/mL | 5.00E+06 | 100% (3/3) |
| Influenza B | Nevada | CEID50/mL | 8.00E+06 | 100% (3/3) |
| Enterovirus (e.g. EV68) | Type 71 | TCID50/mL | 1.00E+05 | 100% (3/3) |
| Respiratory syncytial virus | CH93(18)-18 | TCID50/mL | 1.00E+05 | 100% (3/3) |
| Rhinovirus | A16 | TCID50/mL | 1.00E+05 | 100% (3/3) |
| Chlamydia pneumoniae | VR-1310 | cfu/ml | 6.25E+05 | 100% (3/3) |
| Haemophilus influenzae | Type b; Eagan | cfu/ml | 1.20E+07 | 100% (3/3) |
| Legionella longbeachae | Long Beach 4 | cfu/ml | 9.65E+07 | 100% (3/3) |
| Mycobacterium tuberculosis | H37Ra-1 | cfu/ml | 3.62E+07 | 100% (3/3) |
| Streptococcus pneumoniae | 19F; Z022 | cfu/ml | 2.09E+07 | 100% (3/3) |
| Streptococcus pyogenes | BAA946 | cfu/ml | 7.15E+07 | 100% (3/3) |
| Bordetella pertussis | A639 | cfu/ml | 4.22E+07 | 100% (3/3) |
| Mycoplasma pneumoniae | M129 | CCU/ml | 2.81E+06 | 100% (3/3) |
| Pneumocystis jirovecii (PJP) | W303-Pji | cfu/ml | 7.80E+06 | 100% (3/3) |
| Candida albicans | Z006 | cfu/ml | 9.80E+06 | 100% (3/3) |
| Pseudomonas aeruginosa | Z139; VIM-1 | cfu/ml | 6.05E+06 | 100% (3/3) |
| Staphylococcus epidermis | MRSE; RP62A | cfu/ml | 3.24E+08 | 100% (3/3) |
| Staphylococcus saprophyticus | Z170 | cfu/ml | 9.50E+06 | 100% (3/3) |
| Human coronavirus 229E | ATCC® VR-740DQ | genome copies/µL | 1.30E+04 | 100% (3/3) |
| Human coronavirus OC43 | ATCC® VR-1558D | ng/ul | 2.50E-03 | 100% (3/3) |
| Human coronavirus HKU1 | ATCC® VR-3262SD | genome copies/µL | 2.85E+04 | 100% (3/3) |
| Human coronavirus NL63 | ATCC® VR-3263SD | genome copies/µL | 3.40E+04 | 100% (3/3) |
| SARS-coronavirus | 2003-00592 | cfu/ml | NA* | 100% (3/3) |
| MERS-coronavirus | EMC/2012 | genome copies/µL | NA* | 100% (3/3) |
| Escherichia coli | Clinical Isolate | cfu/ml | 1.92E+08 | 100% (3/3) |
| Burkholderia cepacia | Z066 | cfu/ml | 2.07E+08 | 100% (3/3) |
| Klebsiella pneumoniae | Z148, OXA-48, CTX-M | cfu/ml | 4.15E+08 | 100% (3/3) |
*Information currently not available from supplier
Analytical Specificity – Interfering SubstancesTo assess substances with the potential to interfere with the performance of the Accola SARS-CoV-2 Test, contrived samples with SARS-CoV-2 RNA (SARS-CoV-2 RNA/strain USA_WA1/2020) were tested in replicates of three (3) with each interfering substance at the “worst-case” concentration, and negative samples without RNA were tested in replicates of two (2) with each interfering substance at the “worst-case” concentration. The SARS-CoV-2 RNA was tested at 3X the LoD confirmed in the Limit of Detection Study described above. For each positive sample, RNA was diluted into a pooled negative nasal and throat mix swab matrix to achieve a 3X LoD concentration.The SARS-CoV-2 RNA was tested with an interference concentration representing the highest concentration likely to be found in a respiratory or throat sample. Potentially interfering substances that were sourced in their solid phase were resuspended and diluted to a concentration deemed to be likely worst case. Liquid phase potential interferents were not diluted before testing. Additionally, the SARS-CoV-2 RNA was tested without the interfering substance as a control. Potential interferents and their concentrations, samples tested, and test results are summarized in the table below. No interference was observed with any of the substances tested.
Accula SARS-CoV-2 Interfering Substances Evaluation
| PotentialInterferent | Active Ingredient | Final Concentration | Target | % Agreement withExpected Results |
| Blood (Human) | NA | 25′, | Positive SARS-COV-2(3X LoD) | 100% (313) |
| Negative | 100% (2/2) | |||
| Chloraseptic Max | Phenol 1.5%, Glycerin 33% | Neat | Positive SARS-COV-2(3X LoD)100% (3/3) | |
| Negative | 100% (2/2) | |||
| Cold&Flu ReliefCough Syrup | Acetaminophen 21.7 mg/mL.Dextromethorphan 0.67mg/mL, Guaifenesin 13.3mg/mL. Phenylephrine 0.33mq/mL | Neat | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Listerine Cool MintAntiseptic MouthWash | Eucalyptol 0.092%, Menthol0.042%, Methyl Salicylate0.060%. Thymol 0.064% | Neat | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Cepacol (throatlozenge) | Benzocaine, Menthol | 1 Lozenge/5 mL | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Secrets | Dyclonine Hydrochloride.Menthol | 1 Lozenge/5 mL | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Crest Pro-HealthFluorideToothpaste | Stannous Fluoride 0.454%(0.14% WN Fluoride Ion) | Neat | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Eucalyptus Oil’ | Eucalyptus Oil | Neat | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Advil Liqui-Gels | Ibuprofen | Neat | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Miralax | Polyethylene Glycol | 0.304 g/mL | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Turns ExtraStrength | Calcium Carbonate | 1 tum/2.5 mL | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Food Dye | N/A | Neat | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Whole Milk (Dairy)’ | N/A | 2.% | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| 13.% | Negative | 100% (2/2) | ||
| Orange Juice | N/A | 50% | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Penicillin G | Penicillin G Sodium Salt | 100 mg/mL | Positive SARS-COV-2(3X LoD) 100% (3/3) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Cephalexin | Cephalexin | 25 mg/mL | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) |
| Mucin, Type II(from porcinestomach)2 | Purified mucin protein | 50 mg/mL | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| 100 mg/mL | Negative | 100% (2/2) | ||
| Tobramycin(antibacterial)3 | Tobramycin | 75 mg/mL | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Amoxicillin’ | Amoxicillin | 100 mg/mL | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Anti-viral drug | Zanamivir | 10 mg/mL | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Nasal spray | Phenylephrine | Neat | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Nasal spray | Oxymetazoline | Neat | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Nasal spray | Sodium Chloride | Neat | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| NasalCorticosteroid | Triamcinolone | Neat | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| Zicam (Nasal Gel,homeopathicallergy relief) | Oxymetazoline hydrochloride0.05% | Neat | Positive SARS-COV-2(3X LoD) | 100% (3/3) |
| Negative | 100% (2/2) | |||
| 1 — Milk inhibited target RNA at 12.5%. Milk was diluted to 6.25%, and still inhibited 3 of 3 reactions, and was subsequently diluted to 1.6%, which showed 100% positive target lines. Internal positive control was active and visible at 12.5% milk.2 — Mucin inhibited target RNA at 100 mg/mL. Mucin was diluted to 50 mg/mL and showed 3 positives out of 3 attempts (100%). Internal positive control was active and visible at 100 mg/mL of Mucin.3 — Tobramycin and Amoxicillin positive samples were initially tested without RNA by mistake. These were repeated and yielded 100% expected results.4 — Eucalyptus Oil was used as a substitute for Halls’s Triple Soothing Cough Drops. |
FDA SARS-CoV-2 Reference Panel TestingThe evaluation of sensitivity and MERS-CoV cross-reactivity was performed using reference material (T1), blinded samples, and a standard protocol provided by the FDA. The study included a range-finding study and a confirmatory study for LoD. Blinded sample testing was used to establish specificity and to corroborate the LoD. Testing was performed using the Accula SARS-CoV-2 Test with the Actual Dock. The results are summarized in the table below.
Summary of LoD Confirmation Result using the FDA SARS-CoV-2 Reference Panel
| Reference Materials Provided by FDA | Specimen Type | Product LoD | Cross-Reactivity |
| SARS-CoV-2 | Nasal swab | 4.75 x 102 NDU/mL | N/A |
| MERS-CoV | N/A | ND |
NDU/mL = RNA NAAT detectable units/mLN/A: Not applicableND: Not detected
Clinical EvaluationTesting of Contrived SamplesThirty (30) negative samples and 30 positive contrived samples were tested with the Accula SARS-CoV-2 Test. Negative samples were collected from consented healthy volunteers under IRB approval. A throat swab and a nasal swab was collected from a donor and elute together in a vial of Accula SARS-CoV-2 Buffer. Positive samples were prepared from these thirty negative samples. Positive sampleswere spiked with SARS-CoV-2 RNA (SARS-CoV-2 RNA/strain USA_WA1/2020) at the concentrations shown in the table below.Samples were randomized, de-identified and blinded to the testing operator. Sample concentration and test results are summarized in thetable below. 100% agreement was observed with expected results.
Actual SARS-CoV-2 Evaluation with Throat/Nasal Swab Samples
| Sample Concentration | N | Percent (%) Agreement withExpected Results(Observed/Expected) |
| 2x LoD | 20 | 100 (20/20) |
| 5x LoD | 7 | 100 (7/7) |
| 10x LoD | 2 | 100 (2/2) |
| 50x LoD | 1 | 100 (1/1) |
| Negative | 30 | 100 (30/30) |
Testing of Clinical SamplesRetrospective Specimen StudyFifty (50) retrospective clinical specimens, which had already been tested with a EUA authorized SARS-CoV-2 Real-Time RT-PCR Assay were tested with the Accula SARS-CoV-2 Test. Each specimen was diluted in the minimum amount of Accula SARS-CoV-2 Buffer required to obtain a valid Accula test result (presence of a control line), as VTM can inhibit the assay. Required dilutions ranged from 1:6 to 1:40. Test results are summarized in the table below. One test result was discordant (negative Accula/positive by the comparator). This specimen was re-tested with the Comparator assay and also tested with a second EUA RT-PCR test. The specimen gave negative results in both tests.
| ActualSARS-CoV-2 Test | Comparator Assay | ||
| Positive | Negative | Total | |
| Positive | 23 | 0 | 23 |
| Negative | 26 | 27 | |
| Total | 24 | 26 | 50 |
| Positive Percent Agreement (PPA) | 95.8% (95% CI: 78.88% — 99.89%) | ||
| Negative Percent Agreement (NPA) | 100% (95% CI: 86.77% – 100%) | ||
| Overall Percent Agreement (OPA) | 98.0% (95% CI: 89.35% – 99.95%) |
*1 negative discordant sample was not detected with a secondary comparator test or in re-test with the primary comparator.
Prospective Clinical StudyA prospective study was performed with 52 nasal swabs collected from pediatric patients at a drive-through collection site. Testing was performed with the Accula SARS-CoV-2 test and the comparator method, a EUA authorized RT-PCR SARS-CoV-2 test. Test results aresummarized in the table below.
| ActualSARS-CoV-2 Test | Comparator Assay | ||
| Positive | Negative | Total | |
| Positive | 4 | 0 | 4 |
| Negative | 0 | 48 | 48 |
| Total | 4 | 48 | 52 |
| Positive Percent Agreement (PPA) | 100% (95% CI: 39.76% – 100%) | ||
| Negative Percent Agreement (NPA) | 100% (95% CI: 92.60% – 100%) | ||
| Overall Percent Agreement (OPA) | 100% (95% CI: 93.15% – 100%) |
ASSISTANCE AND CONTACT INFORMATIONFor technical questions or assistance, or if the Accula Dock and/or Accola SARS-CoV-2 Test is not performing as expected, please contact Mesa Biotech at [email protected] or (858) 800-4929.
  |
This symbol indicates that the product is for single-use only. It is not to be re-used |
  |
This symbol indicates that you should consult the instructions for use. |
  |
This symbol is used for both warnings and cautions.A warning indicates the risk of personal injury or loss of life if operating procedures and practices are not correctly followed.A caution indicates the possibility of a loss of data or damage to, or destruction of, equipment if operating procedures and practices are not strictly observed. |
| This symbol indicates that the product has a temperature limitation | |
| This symbol indicates the use-by date | |
  |
This symbol indicates the product batch code. |
  |
This symbol indicates the name and location of the product manufacturer. |
  |
This symbol indicates the product’s catalog number. |
  |
This symbol indicates a positive control. |
| This symbol indicates negative control. | |
  |
Contents sufficient for <n> tests |
| Caution: Federal Law restricts this device to sale by or on the order of a licensed practitioner. | |
| Date | |
  |
For In Vitro Diagnostic Use |
  |
Biohazard: Follow proper infection control guidelines for handling all samples, Test Cassettes, SARS-CoV-2Buffer, and swabs.Properly dispose of all contaminated waste accordingto federal, state, and local requirements. |
This product may be covered by one or more U.S. and/or foreign patents or pending patent applications. See www.mesabiotech.com/patents for details.


Mesa Biotech, Inc.6190 Cornerstone Court EastSuite 220San Diego, CA 92121 USA858-800-4929[email protected]Accula™ SARS-CoV-2 Instructions for UseThe Mesa Biotech logo and Accula™ name are trademarks of Mesa Biotech60061-7 (2021-01) Accula SARS-CoV-2 IFU
References
[xyz-ips snippet=”download-snippet”]

