INSTRUCTIONS FOR USESherlockTM CRISPR SARS-CoV-2 kit Rx OnlyFor Emergency Use Authorization (EUA) only
Rx OnlyFor Emergency Use Authorization (EUA) only
| CATALOG NUMBER | COMPANY |
| TBD | Sherlock Biosciences, Inc.Smart Labs 40 Guest Street, 3 RDFloor Boston, MA 02135, USAPhone: (617) 612-8209 |
Intended Use
The SherlockTM CRISPR SARS-CoV-2 kit is intended for the qualitative detection of nucleic acid from SARS-CoV-2 in upper respiratory specimens (such as nasal swabs, nasopharyngeal swabs, oropharyngeal swabs, nasopharyngeal wash/aspirate, or nasal aspirate) and bronchoalveolar lavage specimens from individuals suspected of COVID-19 by their healthcare provider. Testing is limited to laboratories certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a, to perform high complexity tests.Results are for the identification of SARS-CoV-2 RNA. The RNA of the SARS-CoV-2 RNA is generally detectable in respiratory specimens during the acute phase of infection. Positive results are indicative of the presence of SARS-CoV-2 RNA; clinical correlation with patient history and other diagnostic information is necessary to determine patient infective status. Positive results do not rule out bacterial co-infection with other viruses. The agent detected may not be the definite cause of the disease. Laboratories within the United States and its territories are required to report all positive results to the appropriate public health authoritiesNegative results do not preclude SARS-CoV-2 infection and should not be used as the sole basis for patient management decisions. Negative results must be combined with clinical observations, patient history, and epidemiological information.The Sherlock™ CRISPR SARS-CoV-2 kit is intended for use by qualified clinical laboratory personnel specifically instructed and trained in the techniques of non-automated molecular in vitro diagnostic procedures. The SherlockTM CRISPR SARS-CoV-2 kit is only for use under the Food and Drug Administration’s Emergency Use Authorization.
Summary and Explanation
An outbreak of pneumonia caused by a novel coronavirus (SARS-CoV-2) in Wuhan City, Hubei Province, China was initially reported to WHO on December 31, 2019. On January 31, 2020, Health and Human Services Secretary Alex M. Azar II declared a public health emergency (PHE) for the United States to aid the nation’s healthcare community in responding to SARS-CoV-2. The emergence and rapid spread of SARS-CoV-2 to numerous areas throughout the world, has necessitated preparedness and response in public health laboratories, as well as health care and other areas of society in general. The availability of specific and sensitive assays for the detection of the virus is essential for accurate diagnosis of cases, assessment of the extent of the outbreak, monitoring of intervention strategies, and surveillance studies.
Principles of the Procedure
The Sherlock™ CRISPR SARS-CoV-2 kit has been designed to detect fragments of the Open Reading Frame (ORF1ab, “O”) gene and the Nucleocapsid (“N”) gene of SARS-CoV-2. An included third target is the human RNase P POP7 gene (“RP”) which serves as a control for the extraction of the clinical sample in the absence of a positive SARS-CoV-2 result. A dedicated instrument platform (e.g., thermal cycler) is not required. Amplification can be performed usin™g a heat block, and CRISPR complex activation and reporter cleavage can be run in a standard microplate reader capable of fluorescence detection. The entire reaction from RT-LAMP amplification to CRISPR-based detection of the target analytes can be performed in approximately one hour.The Sherlock™ CRISPR SARS-CoV-2 kit is designed to detect RNA from upper respiratory specimens (such as nasal swabs, nasopharyngeal swabs, oropharyngeal swabs, nasopharyngeal wash/aspirate or nasal aspirate) and bronchoalveolar lavage specimens from patients suspected of COVID-19 by their healthcare provider. RNA is extracted from clinical samples using the PureLinkTM Viral RNA/DNA Mini Kit.The Sherlock CRISPR SARS-CoV-2 kit comprises two steps. Step one is a reverse transcriptase loop-mediated amplification (RT-LAMP) where targeted SARS-CoV-2 genomic RNA is reverse transcribed to DNA, and this DNA is amplified by a strand-displacing DNA polymerase. Step two is the transcription of the amplified DNA to activate the collateral cleavage activity of a CRISPR complex programmed to the target RNA sequence. This collateral activity results in the cleavage of nucleic acid reporters, resulting in a fluorescent readout detected by a plate reader.
Components and Storage
Materials Required (Provided): Each SherlockTM CRISPR SARS-CoV-2 kit consists of the following components:
Table 1: Components Included in Kit
| CapColor | Component | Description | Pack Size –33 Tests perkit | Volume perVial |
| Blue | RP — 10x Primer | RNaseP POP7 gene LAMP Primer Mix | 1 vial | 100 µL |
| N —10x Primer | N gene LAMP Primer Mix | 1 vial | 100 μL | |
| 0 —10x Primer | ORF1ab gene LAMP Primer Mix | 1 vial | 100 μL | |
| 2x RT-LAMP Mix | 2x Warm Start RT-LAMP Mix | 1 vial | 1.1 mL | |
| Yellow | RP — crRNA | RNase P POP7 gene crRNA, 1 μM | 1 vial | 40 μL |
| N — crRNA | N gene crRNA, 1 μM | 1 vial | 40 μL | |
| 0 — crRNA | ORFlab gene crRNA, 1 μM | 1 vial | 40 μL | |
| Reporter | RNase Alert, 2 μM | 1 vial | 200 μL | |
| Red | CRISPR-Casenzyme | LwaCasl3a Enzyme, 500 ng/μL | 1 vial | 50 μL |
| T7 RNAPolymerase | T7 Polymerase, 50 Uha | 1 vial | 70 μLL | |
| rNTP Mix | rNTP Mix, 25 mM of each rNTP | 1 vial | 120 μL | |
| Rnase Inhibitor | Murine RNase Inhibitor, 40 Writ | 1 vial | 75 μL | |
| MgCl2 | 1 M MgCl2 | 1 vial | 50 μL |
Storage and Handling of Kit Components
- The Sherlock™ CRISPR SARS-CoV-2 kit is shipped on dry ice. The components of the kit should arrive frozen. If one or more of the components are not frozen upon receipt or are compromised during shipment, contact Sherlock Biosciences for assistance.
- Store all components at or below -20°C to prevent degradation of reagents.
- Based on individual component shelf life, the approximate shelf life of the kit is estimated to be 12 months.
- Always check the expiration date prior to use. Do not use expired reagents.
- Always work with Sherlock™ CRISPR SARS-CoV-2 kit components on ice.
Materials Required (But Not Provided)
Control Materials
- Positive Control: Quantified extracted SARS-CoV-2 genomic RNA (recommended supplier below)
- Negative Control: Molecular grade, nuclease-free water (recommended supplier below)
Table 2: Control Materials
| Control | Supplier | Part Number | Description |
| SARS-CoV-2Positive Control(SPC) | BEI ResourcesATCC® | NR-52285VR1986D™ | Genomic RNA from SARS-Related Coronavirus 2, Isolate USA-WA1/2020 or equivalent (BEI NR-52285, or ATCC®VR1986D™), diluted to a concentration of 4800 copies/µL |
| NegativeTemplate Control(NTC) | Qiagen | 1039480 | Nuclease-free water for use in Any Molecular Biology application |
RNA Extraction System
Table 3: RNA Extraction System Extraction System
| Extraction System | Manufacturer | Catalog No. |
| PureLink™ Viral RNA/DNA Mini Kit | Thermo Fisher Scientific | 12280050 (50 extractions) |
Equipment
Note: Prior to using, ensure that instruments and equipment have been maintained and calibrated according to the manufacturer’s recommendations.
Table 4: Equipment Equipment
| Equipment | Manufacturer | Model |
| Plate Reader running Gen5 3.08 software | BioTek | NEO2 |
| PC running Microsoft Excel | NA | NA |
Table 5: Additional Equipment and Consumables
| Equipment and Consumables |
| 0.2 mL strip tubes |
| 1.5 mL snap cap tubes, low bind and nuclease-free |
| Molecular grade water (nuclease-free) |
| Dedicated adjustable P-10, P-20, P-100, P-200, and P-1000 pipettes for sample preparation |
| Dedicated adjustable P-10 or P-20 for dispensing template RNA |
| Dedicated adjustable P-10, P-20, P-100, P-200, and P-1000 pipettes for preparing and dispensingmaster mix |
| Dedicated adjustable M-10 and M-100 multichannel pipettes for transferring RT-LAMP amplifiedproduct and CRISPR Cas detection reaction |
| Dedicated electronic pipettes for dispensing master mixes (OPTIONAL) |
| Aerosol barrier tips |
| 384 Corning Black Clear Bottom Low Volume Plate |
| Plate Optical Seal |
| Biosafety Cabinet Class II, for the extraction |
| PCR Workstations, for each portion of the essay set up |
| Heat block with a heated lid capable of maintaining 61°C or PCR instrument with a heated lid |
| Vortex |
| Microcentrifuge |
| Cold blocks or ice |
| Tube racks |
| 8 strip tube opener** |
| Dry Bath/Heat Block* |
| Tabletop Centrifuge* |
| Serological Pipette* |
*required for RNA Extraction only**recommended to reduce contamination risk
Table 6: Additional Supplies
| Dedicated laboratory coat for each area | Powder-free latex, vinyl or nitrile gloves |
| Disposable booties | 20% (v/v) bleach solution (2.0% w/v sodiumhypochlorite in water) |
| Biohazard bag for tip and tube disposal | 70% ethanol |
Warnings and Precautions
- For in vitro diagnostic use (IVD).
- For use under an Emergency Use Authorization (EUA) only.
- For prescription use only.
- Laboratories within the United States and its territories are required to report all positive results to the appropriate public health authorities.
- Follow standard precautions. All patient specimens and positive controls should be considered potentially infectious and handled accordingly.
- Proper personal protective equipment including lab coats, gowns, gloves, eye protection, and a biological safety cabinet are recommended for manipulation of clinical specimens. Refer to Biosafety in Microbiological and Biomedical Laboratories (BMBL) 5th Edition – CDC.
- Do not eat, drink, smoke, apply cosmetics or handle contact lenses in areas where reagents and human specimens are handled.
- Handle all specimens as if infectious using safe laboratory procedures. Refer to Interim Laboratory Biosafety Guidelines for Handling and Processing Specimens Associated with SARS-CoV-2https://www.cdc.gov/coronavirus/2019-nCoV/lab-biosafety-uidelines.html.
- Specimen processing should be performed in accordance with national biological safety recommendations.
- If infection with SARS-CoV-2 is suspected based on current clinical and epidemiological screening criteria recommended by public health authorities, specimens should be collected with appropriate infection control precautions.
- Perform all manipulations of human clinical specimens within a Class II (or higher) biological safety cabinet (BSC). Immediately clean up any spill containing potentially infectious material with 0.5-1% (w/v) sodium hypochlorite (20% v/v bleach). Dispose of cleaningmaterials in a biohazard waste stockpot.
- Report the incident to the supervisor and consult a physician immediately in the event that infectious materials are ingested or come into contact with mucus membranes, open lacerations, lesions or other breaks in the skin.
- The use of non-recommended reagent volumes may result in a loss of performance and may also decrease the reliability of the test results.
- The use of non-recommended volumes and concentrations of the RNA/ DNA sample may result in a loss of performance and may also decrease the reliability of the test results.
- The use of non-recommended consumables with instruments may adversely affect test results.
- Do not mix reagents from different lots.
- RNA should be maintained on a cold block or ice during preparation and use to ensure stability.
- Primers, CRISPR guide RNA (crRNA) stocks (including aliquots), enzymes, and RT-LAMP amplification master mix must be thawed and maintained on a cold block at all times during preparation and use.
- Return all components to the appropriate storage condition after preparing the working reagents.
- Workflow in the laboratory should proceed in a unidirectional manner.
- Amplification technologies are sensitive to the accidental introduction of products from previous amplification reactions. Incorrect results could occur if either the clinical specimen or reagents used in the amplification step become contaminated by the accidental introduction of the amplification product (amplicon).
- Maintain separate areas for assay setup and handling of nucleic acids.
- Always check the expiration date prior to use. Do not use expired reagents. Do not substitute or mix reagent from different kit lots or from other manufacturers.
- Change aerosol barrier pipette tips between all manual liquid transfers.
- During the preparation of samples, compliance with good laboratory techniques is essential to minimize the risk of cross-contamination between samples, and the inadvertent introduction of nucleases into samples during and after the extraction procedure.A proper aseptic technique should always be used when working with nucleic acids.
- Maintain separate, dedicated equipment (e.g., pipettes, microcentrifuges) and supplies (e.g., microcentrifuge tubes, pipette tips) for assay setup, handling of extracted nucleic acids, and handling post-amplification products.
- Perform work in a unidirectional workflow in separate locations, from areas without specimen/nucleic acid or amplicon to areas with amplified nucleic acid
- Wear a clean lab coat and powder-free disposable gloves (not previously worn) when setting up assays.
- Change gloves between samples and whenever contamination is suspected. Change gloves after tubes containing amplified products are handled before touching other tubes, equipment, etc.
- Keep reagent and reaction tubes capped or covered as much as possible.
- Do not interchange vial or bottle caps, as cross-contamination may occur.
- Work surfaces, pipettes, and centrifuges should be cleaned and decontaminated with cleaning products such as 20% bleach and “RNase AWAY® ” to minimize the risk of nucleic acid or RNase contamination. Residual cleaning solutions should be removed using 70% ethanol.
- Dispose of unused kit reagents and human specimens according to local, state, and federal regulations.The product contains no substances which at their given concentration, are considered to be hazardous to health or the environment.
The product contains no substances which at their given concentration, are considered to be hazardous to health or the environment.
HMIS
| Health | 0 |
| Flammability | 0 |
| Reactivity | 0 |
Specimen Collection, Handling, and Storage
Adequate, appropriate specimen collection, storage, and transport are important in order to obtain accurate test results. Training in correct specimen collection procedures is highly recommended to assure good quality specimens and results. CLSI MM13-A may be referenced as an appropriate resource.
Collecting the Specimen
- Refer to Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Patients Under Investigation (PUIs) for 2019 Novel Coronavirus (2019-nCoV) https://www.cdc.gov/coronavirus/2019-nCoV/guidelines-clinical-specimens.html
- A sample collection device is not a part of the assay kit. Follow specimen collection device manufacturer instructions for proper collection methods.
- Swab specimens should be collected using only swabs with a synthetic tip, such as nylon or Dacron®, and an aluminum or plastic shaft. Calcium alginate swabs are unacceptable and cotton swabs with wooden shafts are not recommended. Place swabs immediately into sterile tubes containing 2-3 ml of viral transport media (i.e. VTM, UTM, M4RT).
Transporting Specimens
- It is the shipper’s responsibility to ensure that appropriate shipping materials are used. Please refer to IATA and local regulations.
- Refrigerate specimens at 2-8°C and ship overnight on dry ice.
Storing Specimens
It is recommended that specimens be kept at -20°C for up to 7 days. For storage longer than 7 days, specimens should be frozen at -70°C. Repeated freezing and thawing of a specimen should be avoided. If a specimen is kept for retesting, it should be aliquoted in different tubes to avoid freezing and thawing cycles. The temperature in the storage areas should be monitored and recorded regularly to identify potential fluctuations. Domestic refrigerators/ freezers with wide temperature fluctuations are not suitable for the storage of frozen specimens (CDC, 2020).
Reagent Controls and Preparation
SARS-CoV-2 Positive Control (spc) Preparation: · Precautions: This reagent should be handled with caution to prevent possible contamination.Freeze-thaw cycles should be avoided. Maintain on the cold block when thawed.
- Dilute the SPC with nuclease-free water to achieve the working concentration of the SPC (4,800 gene copies/µL). Make single-use aliquots (approximately 20 µL each) and store them at -70oC.
- Thaw a single aliquot of the positive control for each experiment and keep on the cold block until adding to the RT-LAMP reaction. Discard any unused portion of the aliquot.
No Template Control (NTC)
- Nuclease-free water
Quality Control
- Quality control requirements must be performed in conformance with local, state, and federal regulations or accreditation requirements and the user’s laboratory’s standard quality control procedures. For further guidance on appropriate quality control practices, refer to 42 CFR 493.1256.
- Quality control procedures are intended to monitor reagent and assay performance.
- Always include a negative control (NTC) for all three SherlockTM SARS-CoV-2 kit targets (O, N, and RP), and positive control (SPC) for each of the SARS-CoV-2 specific targets (O and N) in each amplification and detection run.
- All clinical samples must be tested for the presence of the human RNase P (RP) target to control for specimen quality and extraction, which acts as the internal control.
Nucleic Acid Extraction and Assay Set up
All procedures should be performed in a BSL2 laboratory, and specimens should be handled within a Biological Safety Cabinet. All necessary safety precautions should be taken according to the Laboratory guidelines. Precautions must also be taken to prevent cross-contamination of samples.
Separate work areas should be used for:
- Nucleic acid extraction
- Reagent preparation (e.g. preparation of master mixes; NO amplified reactions, target solutions, or clinical specimens should be brought into this area. After working in this area, laboratory coat and gloves should be changed before moving into the nucleic acid addition area)
- Nucleic acid addition
- RT-LAMP Amplification (e.g. thermocyclers/heat blocks)
- Post-amplification detection (After working in this area, laboratory coat and gloves should be changed and disposed of)
General Handling
- Proper microbiological, aseptic techniques should always be used when working with RNA. Hands and dust particles may carry bacteria and molds and are the most common sources of RNase contamination.
- Always wear powder-free latex, vinyl, or nitrile gloves while handling reagents, tubes and RNA samples to prevent RNase contamination from the surface of the skin or from dusty laboratory equipment.
- Change gloves frequently and keep tubes closed when not in use.
- During the procedure, avoid delays and keep everything on cold blocks of ice when possible to avoid degradation of RNA by endogenous or residual RNases.
- Clean working surfaces, pipettes, and equipment with 20% bleach or other solution that can destroy nucleic acids and RNases. To eliminate accelerated deterioration of any plastics and metals, wipe down with 70% ethanol after using 20% bleach.
- Make sure all bleach is removed to eliminate possible chemical reactions between bleach and guanidine thiocyanate which is present in the extraction reagents.
PROCEDURE
- Nucleic Acid Extractiona. The SherlockT™ SARS-CoV-2 kit uses the PureLinkTM Viral RNA/DNA Mini Kit (Cat. 12280050). Add 200 L of sample to 225 L of pre-aliquoted Lysis Buffer/proteinase K mixture (total input sample lysate volume is 425 L). Proceed with the extraction as indicated in the Instructions For Use for the kit with the addition of one step.Note: Additional Nucleic Acid Extraction step required. Immediately prior to RNA elution, open the assembly lid and air-dry the column membrane for 15 minutes at room temperature before eluting with 30 L of nuclease-free water.b. Up to 8 µL of the extracted RNA can be used per reaction. Extracted RNA is extremely sensitive to degradation caused by RNases, follow general laboratory precautions for handling RNA. Store extracted RNA <-70°C if not using immediately.
- RT-LAMP Master Mix Preparationa. Label a new 1.5 mL microcentrifuge tube for each target (O, N, and RP) and prepare an RT-LAMP Master Mix consisting of the 2x RT-LAMP Mix and the appropriate 10x Primer Mix using the recipe in Table 6 below. Make enough of each master mix for all samples to be tested and the necessary controls for each run.b. Pulse vortex RT-LAMP Master Mix for 3 seconds and spin down for 3 seconds in a microcentrifuge after all components are added.Table 7: Target Specific RT-LAMP Master Mix Recipe
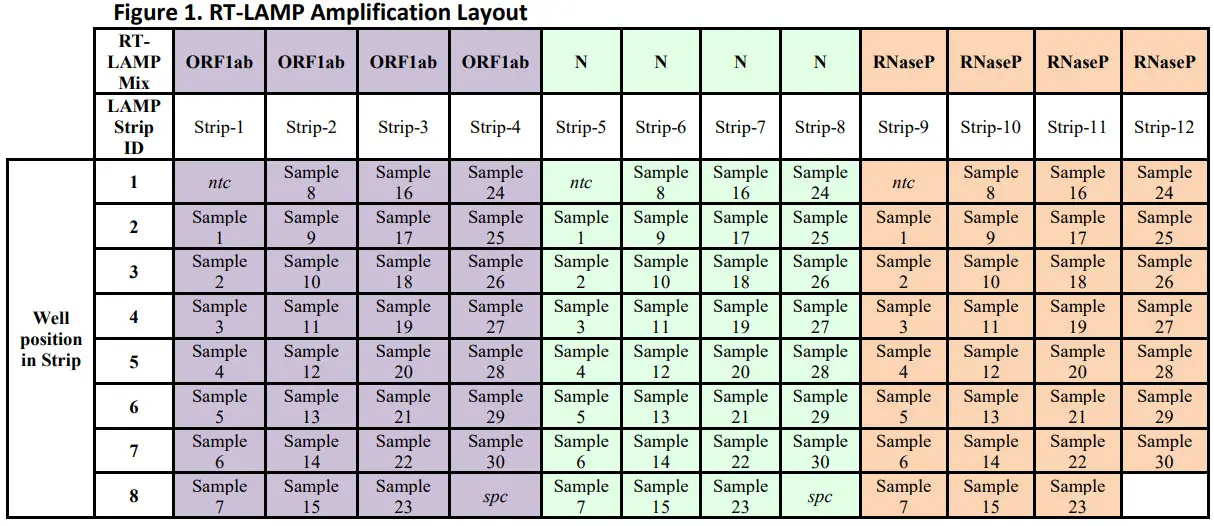
Reagent Cap Color Volume per reaction Volume total 2x RT-LAMP Mix Blue 10 µL 10 µL x (N +1) 10x Primer (N, O, or RP) Blue 2 µL 2 µL x (N + 1) Total Volume NA 12 µL 12 µL x (N +1) - RT-LAMP Amplificationa. Label a strip tube (0.2 mL) with the target name (i.e. O) and strip number according to recommended layouts in Figure 1. Add 12 µL of the RT-LAMP Master Mix into one well for each sample and control to be amplified. i. Repeat step 3.a. for the remaining 2 targets using a new strip tube for each target (i.e. N, or RP).b. Add 8 µL of extracted RNA in each respective strip tube containing the RT-LAMP Master mix. Vortex the strip tube for 3 seconds and spin down for 3 seconds in microcentrifuge with a 0.2 mL tube adaptor. N = number of extracted samples plus a number of controls. Prepare enough for 1 extra (N + 1) sample to allow for overage during reaction set-up.Table 8: RT-LAMP Assay Components and reaction volume
Reagent Volume per reaction RT-LAMP Master Mix 12 µL RNA Sample or Controls 8 µL Total Volume 20 µL c. Incubate the strip tubes on the thermocycler/heating block set to 61°C for 40 minutes.Note heat the lid to 99°C for the incubation. Pulse spin the tubes after the 40-minute incubation.
- CRISPR Cas Reaction Preparationa. Preheat a fluorescence microplate reader to 37°C.b. For each target tested label a 1.5 mL tube with the target name (i.e. O) and “Cas Mix”. Prepare a CRISPR Cas Master Mix using the following recipe in Table 8 below, scaling as required for the number of assays to be run (one Cas assay for every RT-LAMP reaction). A minimum total volume to complete 5 reactions is recommended (N 5 in Table 8).Note: The mix, without MgCl2, can be prepared while the RT-LAMP amplification is running and stored in a cool block. Add the MgCl2 to the mix last, immediately before moving on to the next step.i. Repeat step 5.b. for the remaining 2 targets using a new 1.5 mL tube for each target (i.e. N, or RP).c. Pulse vortex for 3 seconds and spin down for 3 seconds in a microcentrifuge after all components are added.CAUTION: Do not allow the completed mix to sit for longer than 10 minutes prior to moving on to the next step.Table 9: Target CRISPR Cas Master Mix Recipe
Reagent Cap Color Volume per Reaction Volume total Reporter Yellow 1.56 µL 1.56 µL x (N + 1) crRNA(N or O or RP) Yellow 0.56 µL 0.56 µL x (N + 1) rNTP Mix Red 1 µL 1 µL x (N + 1) T7 RNA Polymerase Red 0.5 µL 0.5 µL x (N + 1) RNase Inhibitor Red 0.63 µL 0.63 µL x (N + 1) CRISPR-Cas enzyme Red 0.32 µL 0.32 µL x (N + 1) Water n/a 15.2 µL 15.2 µL x (N + 1) MgCl2 Red 0.23 µL 0.23 µL x (N + 1) Total Volume NA 20 µL 20 µL x (N + 1) N = number of extracted samples plus a number of controls. Prepare enough for 1 extra (N + 1) sample to allow for overage during reaction set-up.
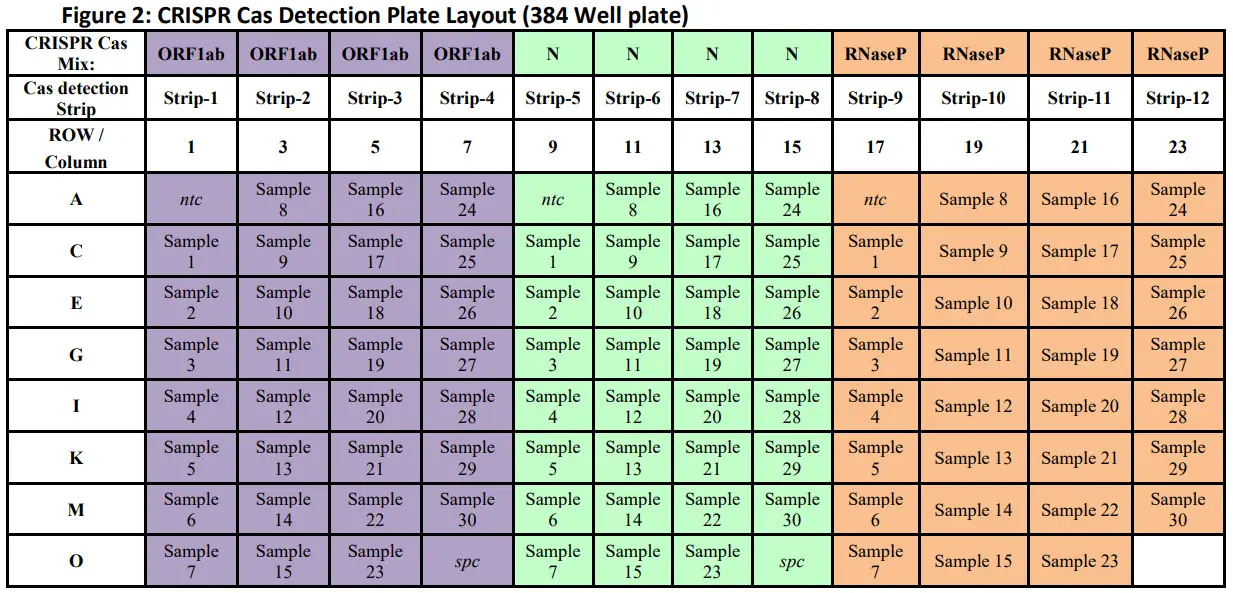
- CRISPR Cas DetectionCaution: Perform work in a unidirectional workflow in separate locations, from areas without specimen/nucleic acid or amplicon to areas with the amplified nucleic acid.a. Label a strip tube (0.2 mL) with the target name (i.e. O) and strip number according to recommend plate layout in Figure 2. Add 20 µL of the CRISPR Cas Master Mix made in step 4 into one well for each sample and control to be amplified. Close strip tube.i. Repeat step 5.a. for the remaining 2 targets using a new strip tube for each target (i.e. N, or RP).b. In an isolated, clean dead box, carefully open one of the aliquoted CRISPR Cas Master Mix strip tubes (i.e. O) using a strip tube opener.c. Open the corresponding target (i.e. O) RT-LAMP amplicon strip tube, placed in a different rack after amplification in step 3.c. is complete. Change gloves.d. Using a multichannel pipette carefully adds 5 µL of the RT-LAMP amplicon to the corresponding CRISPR Cas Master Mix strip tube. Eject tips.Table 10: CRISPR Cas Detection Components and reaction volume
Reagent Volume per reaction RT-LAMP Amplicon (N, O, RP) 5 µL CRISPR Cas Master Mix 20 µL Total Volume 25 µL e. Carefully close the caps of the CRISPR Cas Detection strip tube (RT-LAMP amplicon + CRISPR Cas Master Mix). Carefully close the amplified RT-LAMP amplicon strip tubes and dispose after use. Change gloves.f. Flick the CRISPR Cas Detection strip tube to mix and spin down for 3 seconds in a mini centrifuge with a 0.2 mL tube adaptor. Change gloves.i. Repeat step 5.b. to 5.f. for the remaining CRISPR Cas Master Mix strip tubes.g. Carefully open each CRISPR Cas Detection strip tube using a strip tube opener. Change gloves. h. Using a multichannel pipette, carefully add 20 µL from the CRISPR Cas Detection strip tube to a Corning® 384 Well Plate (Black/Clear Bottom) according to recommended plate layout, Figure 2.CAUTION: Do not go to the second stop of the pipette to avoid the introduction of bubbles to the reaction wells.i. Repeat step 5.g. to 5.h. for the remaining CRISPR Cas Detection strip tubes.i. Seal the plate with an Optical Seal.j. Open the plate reader software to create a read procedure.i. Set setpoint temperature to 37°C.ii. Select “Kinetic” run reading with a total read time of 10 minutes, and data collection intervals at 2.5 minutes.iii. Select filter settings in reading details to 485nm/528nm filter set with the gains setting set to “extended”.iv. Highlight the rows and columns of the plate to be read in the plate settings.v. If a warning about “Max V” calculations appears, press “OK” and continue.vi. Press the green arrow to start, (i.e. “Create experiment and read now”).vii. Save the experiment in a designated place with an appropriate unique nameviii. When plate loader, extends, load plateCAUTION: Ensure plate is loaded incorrect orientationix. Press “OK” to load the plate.
- Recommended Layouts


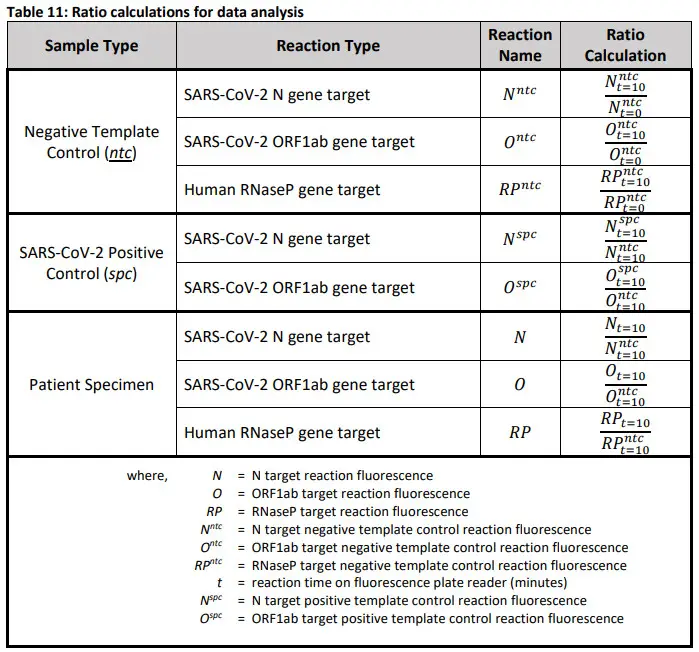
- Data Extraction and Analysisa. After the completion of the plate reader-run, open plate reader software and open experiment file.b. Select and export data for all samples and controls according to manufacturer’s instructions.Export the data from these wells to an excel spreadsheet.c. For the negative template control (“NTC”), SARS-CoV-2 Positive Controls (“SPC”), and patient specimens calculate the ratios in Table 10 below:Table 11: Ratio calculations for data analysis

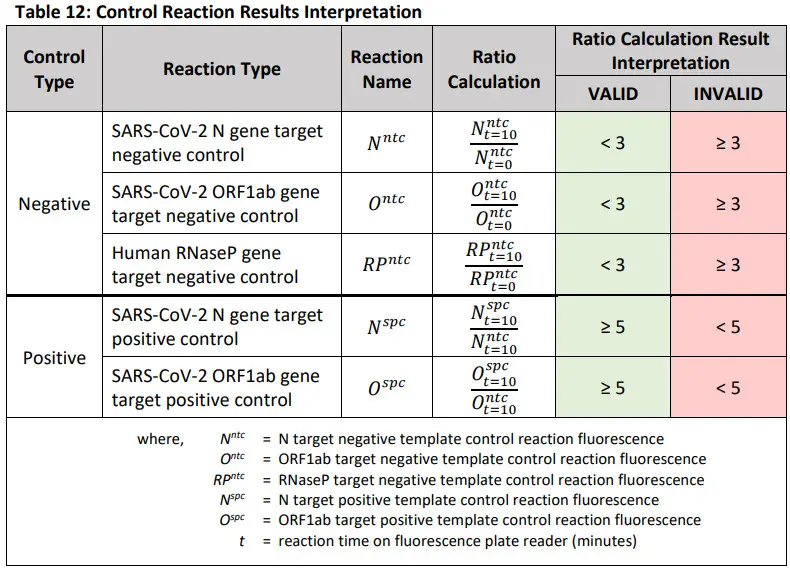
Assay Controls and Interpretation of ResultsAssay ControlsNegative Template Control Reactions (NTC): Negative template control (NTC) reactions are used to monitor reagent and/or environmental contamination. There are three negative control reactions, one for each primer/crRNA set – (i) SARS-CoV-2 N gene, (ii) SARS-CoV-2 ORF1ab gene, and (iii) human RNase P gene. Negative template control reactions are created by substituting the volume of sample material in the RT-LAMP reaction with an equal volume of nuclease-free water.
Positive Control Reactions (spc): Positive control (SPC) reactions are used to monitor gross reagent failures, such as reagent degradation, or incorrect assay set-up. There are two positive control reactions, one for each SARS-CoV-2 target – (i) N gene, and (ii) ORF1ab gene. Positive control reactions are created by substituting the volume of sample material in the RT-LAMP reaction with an equal volume of extracted SARS-CoV-2 viral RNA at a stock concentration of 4800 copies/μl.
CAUTION: If any control reaction is INVALID as defined in the table below, see troubleshooting.

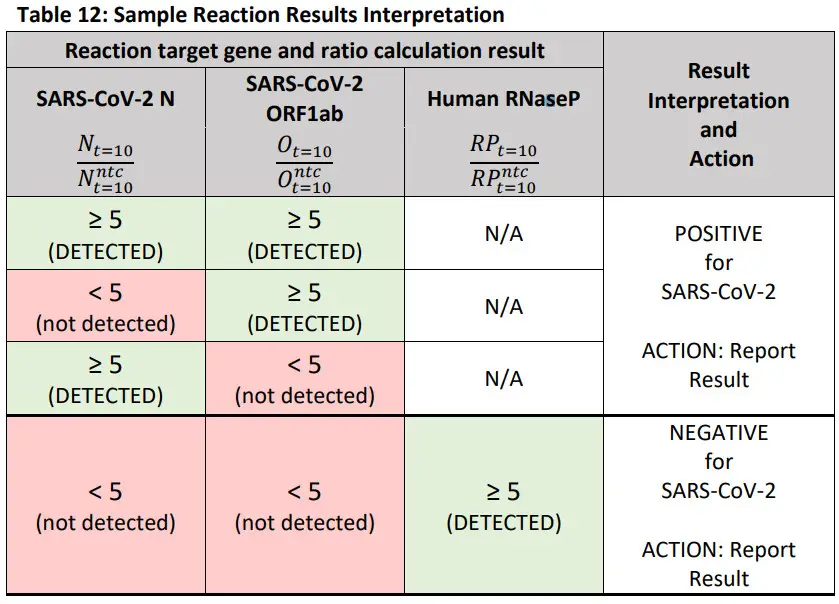
Interpretation of Sample Results
- Unknown clinical sample results should only be assessed once all control reactions have been determined to be valid and acceptable.
- A result is Positive if either SARS-CoV-2 target (N or ORF1ab) reaction has a greater than or equal to 5-fold increase in fluorescence measurement at 10 minutes over the corresponding valid negative template control reaction fluorescence measurement at 10 minutes.
- A result is Negative if both SARS-CoV-2 target (N and ORF1ab) reactions have a less than 5-fold increase in reaction fluorescence measurement at 10 minutes over the corresponding valid negative template control reaction fluorescence measurement at 10 minutes AND the RNaseP target reaction has a greater than or equal to 5-fold increase in fluorescence measurement at 10 minutes over the corresponding valid negative template control reaction fluorescence measurement at 10 minutes.
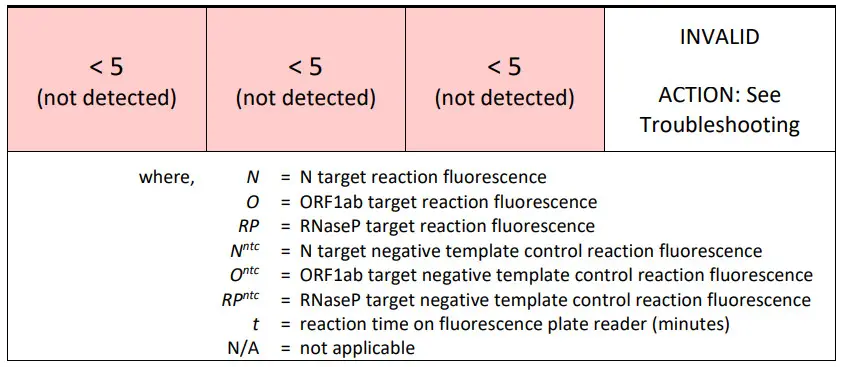
- A result is Invalid if all target (N, ORF1ab, and RNaseP) reactions have a less than 5-fold increase in reaction fluorescence measurement at 10 minutes over the corresponding valid negative template control reaction fluorescence measurement at 10 minutes.
CAUTION: If the sample reaction is INVALID as defined in the table below, see troubleshooting.


Troubleshooting
User Errors
- Good Clinical Laboratory Practices (GCLP) for Molecular Biology Based Tests Used In Diagnostic Laboratories (Viana & Wallis, 2011) are necessary for the use of this product. This product is not intended to be used by untrained personnel. The user needs to have molecular biology experience and be familiar with the proper pipetting technique to prevent errors, such as splashes, crossover contamination, and errors on volume selection.
- Pipette tips must be replaced after every pipetting. Gloves must be replaced often. Equipment must have calibration up to date for the pipettes and thermocyclers, when applicable.
- A 90 minutes online training for Good Laboratory Practices for Molecular Genetics Testing (Centers for Disease Control and Prevention, 2017) is available at the CDC website at the following link:https://www.cdc.gov/labtraining/training-courses/good-lab-practices-molecular-genetics-testing.html Invalid ResultsSARS-CoV-2 Positive Control (SPC) not detected.
Possible causes:
- Pipetting errors (control in wrong well, missing a well, inadequate amount of a reagent)
- Incorrect dilution of positive control nucleic acid
- Incorrect placement of tubes into heat block/PCR machine
- Incorrect placement of plate in plate reader
- Degraded reagents due to incorrect storage temperature
- Use of expired reagents
- Use of incorrect reagent
- If the SARS-CoV-2 Positive Control is invalid, the run should be considered invalid and the user should re-test the samples by re-extraction and use a fresh aliquot of the diluted SARS-CoV-2 Positive Control.
- If the positive control fails again, then an investigation should be conducted to identify possible causes for error, and depending on the investigation results and risks identified in the process, the samples may need to be re-run.
- If failure of the positive control happens a third time after pre-extraction and re-amplification, test with a new lot of Sherlock™ CRISPR SARS-CoV-2 kit reagents.
- If still failing, please contact Sherlock Biosciences.
RNase P (PR) is not being detected in patient samples.
Possible causes:
- Not enough nuclear material in the patient sample
- The extraction was performed incorrectly
- Inhibitors present in a patient sample
- Pipetting errors (control in wrong well, missing a well, inadequate amount of a reagent)
- Incorrect placement of tubes into heat block/PCR machine
- Incorrect placement of plate in a plate reader
- Use of incorrect reagent
- If the RP is invalid, the run should be considered invalid and the user should re-test the samples by re-extraction.
- If the RP fails again, then an investigation should be conducted to identify possible causes for error, and depending on the investigation results and risks identified in the process, the samples may need to be re-run.
- If failure of the RP happens a third time after pre-extraction and re-amplification, test with a new lot of Sherlock™ CRISPR SARS-CoV-2 kit reagents.
- If still failing, please contact Sherlock Biosciences.
Limitations
- All users should be qualified by training or experience to perform molecular diagnostic test procedures.
- Sherlock™ Biosciences will limit the distribution of this device to laboratories that have been certified to perform CLIA high complexity testing.
- The test was validated for use only with upper respiratory specimens.
- Negative results do not preclude SARS-CoV-2 infection and should not be used as the sole basis for treatment or other patient management decisions. Optimum specimen types and timing for peak viral levels during infections caused by SARS-CoV-2 have not been determined. Collection of multiple specimens (types and time points) from the same patient may be necessary to detect the virus.
- A false-negative result may occur if a specimen is improperly collected, transported, or handled.False-negative results may also occur if amplification inhibitors are present in the specimen or if inadequate numbers of organisms are present in the specimen.
- Positive and negative predictive values are highly dependent on prevalence. False-negative test results are more likely when the prevalence of the disease is high. False-positive test results are more likely when prevalence is moderate to low.
- If the virus mutates in the RT-PCR target region, SARS-CoV-2 may not be detected or may be detected less predictably. Inhibitors or other types of interference may produce a false negative result. An interference study evaluating the effect of common cold medications was not performed.
- Test performance can be affected because the epidemiology and pathology of disease caused by SARS-CoV-2 is not fully known. For example, clinicians and laboratories may not know the optimum types of specimens to collect, and when during the course of infection these specimens are most likely to contain levels of virus that can be readily detected.
- Detection of viral RNA may not indicate the presence of an infectious virus or that SARS-CoV- 2 is the causative agent for clinical symptoms.
- The performance of this test has not been established for monitoring the treatment of SARS- CoV-2 infection.
- The performance of this test has not been established for screening of blood or blood products for the presence of SARS-CoV-2.
- This test cannot rule out diseases caused by other bacterial or viral pathogens.
The Sherlock™ CRISPR SARS-CoV-2 kit Letter of Authorization, along with the authorized Fact Sheet for Healthcare Providers, the authorized Fact Sheet for Patients and authorized labeling are available on the FDA website: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19emergency-use-authorizations-medical-devices/vitro-diagnostics-euas
Use of the Sherlock™ CRISPR SARS-CoV-2 kit must follow the procedures outlined in these manufacturer’s Instructions for Use and the conditions of authorization outlined in the Letter of Authorization. Deviations from the procedures outlined are not permitted under the EmergencyUse Authorization (EUA). To assist clinical laboratories running the Sherlock™ CRISPR SARS-CoV-2 kit, the relevant Conditions of Authorization are listed verbatim below and are required to be met by laboratories performing the EUA test.
- Authorized laboratories 1 will include reports of the results of the Sherlock™ CRISPR SARS- CoV-2 kit, the authorized Fact Sheet for Healthcare Providers, and the authorized Fact Sheet for Patients. Under exigent circumstances, other appropriate methods for disseminating these Fact Sheets may be used, which may include mass media.
- Authorized laboratories will perform the Sherlock™ CRISPR SARS-CoV-2 kit as outlined in the Sherlock™ CRISPR SARS-CoV-2 kit Instructions for Use Package Insert. Deviations from the authorized procedures, including authorized extraction methods, authorized clinical specimen types, authorized control materials, authorized other ancillary reagents, and authorized materials required to perform the Sherlock™ CRISPR SARS-CoV-2 kit are not permitted.
- Authorized laboratories that receive your product will notify the relevant public health authorities of their intent to run your product prior to initiating testing.
- Authorized laboratories will have a process in place for reporting test results to healthcare providers and relevant public health authorities, as appropriate.
- Authorized laboratories will collect information on the performance of the test and report to:DMD/OHT7-OIR/OPEQ/CDRH (via email: CDRH-EUA- ) and to Sherlock Biosciences (via phone: 617-702-6263 or via email: [email protected] ) any suspected occurrence of false positive or false negative results and significant deviations from the established performance characteristics of the test of which they become aware.
- All laboratory personnel using the test must be appropriately trained/experienced in molecular in vitro diagnostic test techniques, use appropriate laboratory and personal protective equipment when handling this kit, and use the test in accordance with the authorized labeling.
- Sherlock™ Biosciences and authorized laboratories will ensure that any records associated with this EUA are maintained until otherwise notified by FDA. Such records will be made available to FDA for inspection upon request.
1 The letter of authorization refers to, “United States (U. S.) laboratories certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a, to perform high complexity tests” as “authorized laboratories.”
Performance Characteristics
Analytical Sensitivity (Limit of Detection):The Limit of Detection (LoD) for the Sherlock ™ CRISPR SARS-CoV-2 kit was determined to be the lowest concentration of genomic viral RNA (copies/µL of VTM) at which ≥ 95% of all replicates test positive. As the assay contains two SARS-CoV-2 targets (N and O), the LoD for each target was independently determined and confirmed. The LoD claimed for the kit is the higher of the two values.To determine the LoD, limiting dilutions of quantified extracted genomic RNA were spiked into a clinical matrix composed of pooled nasopharyngeal swabs after the initial lysis step of the PureLink™ Viral RNA/DNA Mini Kit (Thermo Fisher) to prevent the lysis of RNA prior to extraction. After extraction, the samples were processed according to the Sherlock™ SARS-CoV-2 kit workflow. Initially, three (3) replicate ten (10) dilutions were tested. The tentative LoD was determined to be the estimated lowest concentration where 3/3 replicates were positive for a given target. This LoD was confirmed by testing at 1x and 1.5x this concentration in the same sample matrix as above. Results are shown in Table 14 below
Table 14: LoD determination
| Viral Copies inSample (cp/pLVTM) | TotalPositive(ORFlab) | TotalPositive(N) | TotalPositive(RNaseP) |
| 18.0 | 3/3 | 3/3 | 3/3 |
| 11. | 3/3 | 3/3 | 3/3 |
| 7. | 3/3 | 3/3 | 3/3 |
| 6. | 3/3 | 3/3 | 3/3 |
| 5. | 2/3 | 3/3 | 3/3 |
| *4.5 | 3/3 | 3/3 | 3/3 |
| 4. | 1/3 | 3/3 | 3/3 |
| 3. | 3/3 | 3/3 | 3/3 |
| 2. | 1/3 | 3/3 | 3/3 |
| **0.9 | 1/3 | 3/3 | 3/3 |
| 0.0 | 0/3 | 0/3 | 3/3 |
| Positive Control | 3/3 | 3/3 | N/A |
| NegativeControl | 0/3 | 0/3 | 0/3 |
*Estimated LoD of ORF1ab target (4.5 cp/uL VTM)**Estimated LoD of N target (0.9 cp/uL VTM)
The LoD was confirmed by testing at 1x (4.5 cp/µL and 0.9 cp/ µL for ORF1ab and N, respectively) and 1.5x this estimated concentration for each target (6.75 cp/ µL and 1.35 cp/ µL for ORF1ab and N, respectively). The LoD of each target was determined to be the concentration at which ≥19/20 replicates for each assay target was positive. The LoD for the ORF1ab target was determined to be6.75 cp/µL VTM. The LoD for the N target was determined to be 1.35 cp/µL VTM.
Table 15A: LoD confirmation
| Targe | Viral Copies in Sample (cp/uLVTM) | # ofSamples | # ofDetected | DetectionRate (%) |
| ORF1ab | 4.5 | 20 | 17 | 85 |
| ORF1ab | 6.75 | 20 | 19 | 95 |
| N | 0.9 | 20 | 17 | 85 |
| N | 1.35 | 20 | 20 | 100 |
The confirmed LoD for the Sherlock™ CRISPR SARS-CoV-2 kit is 6.75 cp/uL VTM
FDA SARS-CoV-2 Reference Panel Testing
The evaluation of sensitivity and MERS-CoV cross-reactivity was performed using reference material (T1), blinded samples, and a standard protocol provided by the FDA. The study included a range-finding study and a confirmatory study for LoD. Blinded sample testing was used to establish specificity and to confirm the LoD.Testing was performed using the PureLink™ Viral RNA/DNA Mini Kit (Cat. 12280050) extraction method and BioTek NEO2 Plate Reader instrument running Gen5 3.08 software. The results are summarized in Table 15B.
Table 15B: Summary of LoD Confirmation Result using the FDA SARS-CoV-2 Reference Panel
| Reference Materials Provided by FDA | Specimen Type | Product LoD | Cross-Reactivity |
| SARS-CoV-2 | Pooled COVID-19 negativenasopharyngeal swab matrix spiked with a CBER heat inactivatedCOVID-19 strain or MERS-CoV incell culture media | 0.6×10 4 NDU/mL | N/A |
| MERS-CoV | N/A | ND |
NDU/mL = RNA NAAT detectable units/mLN/A: Not applicableND: Not detected
Analytical Inclusivity:In silico analysis was performed to determine the reactivity of the SHERLOCK™ CRISPR SARS-CoV-2 kit. All (9166 individual strains) SARS-CoV-2 genomes available from GISAID on 04/15/2020 and defined as complete by GISAID (>29,000 bp) were downloaded. The 9166 genomes were aligned with all primer and crRNA binding regions for both ORF1ab and N targets, strains with mismatches were identified, and the dataset was refined by removing samples from animals (e.g. pangolin, bat) and ambiguous sequencing data (e.g. N’s). N=109 (1.19%) genomes were identified with a single mismatch to the ORF1ab target sequences. N=628 (6.85%) genomes were identified with a single mismatch (627 genomes) or double mismatch (1 genome) to the N target sequences. No genome was identified with a mismatch in both the ORF1ab and N targets.
It is therefore determined that all available SARS-CoV-2 target analyte sequences will be detected by at leastone of the SHERLOCK ™ CRISPR SARS-CoV-2 kit target analyte tests.Analytical Specificity:All primers and crRNAs were analyzed with BLASTn using public domain nucleotide databases and excluding results with the taxonomy ID for SARS-CoV-2. The next database was used as described above for general exclusivity testing except this database was also filtered to only include sequences for taxonomy identifiers (taxid) of the high priority organisms (see Table 15 below). Search parameters were automatically adjusted for short input sequences. Additional settings included: expect threshold 1000, match score 1, and mismatchscores -3 and penalty to create a gap in an alignment 5 and extend a gap in an alignment 2.
Significant homology (>80%) was identified between Bat SARS and a single primer in the ORF1ab target.Significant homology (>80%) was identified between Bat SARS, SARS, and Influenza B and a single primer or guide in the N target. Since LAMP amplicon generation is not possible with a single primer there is little risk of cross-reactivity with the organisms.
Of the organisms identified with high homology, microbial interference is unlikely from Bat SARS or SARS because they are not common human pathogens. Microbial interference from Influenza B is unlikely because there are 2 mismatches at the terminal 3’ end of the primer.In summary, the ORF1ab and N primers and crRNAs designed for the specific detection of SARS-CoV-2 showed no significant homologies that are likely to cause cross-reactivity or microbial interference with the human genome and transcriptome or with other organisms including bacteria and viruses.
Table 16 – List of High Priority Organisms for Specificity / Exclusivity Assessment
| Other high priority pathogens from thesame genetic family | High priority organisms likely in the circulating area |
| Human coronavirus 229E | Adenovirus (e.g. C1 Ad. 71) |
| Human coronavirus OC43 | Human Metapneumovirus (hMPV) |
| Human coronavirus HKU1 | Parainfluenza virus 1-4 |
| Human coronavirus NL63 | Influenza A & B |
| SARS-coronavirus | Enterovirus (e.g. EV68) |
| MERS-coronavirus | Respiratory syncytial virus |
| Rhinovirus | |
| Chlamydia pneumoniae | |
| Haemophilus influenzae | |
| Legionella pneumophila | |
| Mycobacterium tuberculosis | |
| Streptococcus pneumonia | |
| Streptococcus pyogenes | |
| Bordetella pertussis | |
| Mycoplasma pneumonia | |
| Pneumocystis jirovecii (PJP) | |
| Human genome | |
| Candida albicans | |
| Pseudomonas aeruginosa | |
| Staphylococcus epidermis | |
| Staphylococcus salaries |
Wet testing against high-risk pathogenic organisms of the respiratory tract, selected based on disease prevalence, disease risk, homology to assay-specific targets, and homology to the SARS-CoV-2 genome, was performed to confirm the results of the in silico analysis. Each organism identified in Table 17 below was tested in triplicate with the Sherlock™ CRISPR SARS-CoV-2 kit by spiking diluted organism stock into lysistreated pooled nasopharyngeal swab matrix.All replicates were negative for SARS-CoV-2 detection.
Table 17: Wet testing of potential cross-reactive organisms
| Organism | ATCC Cat. Number | Concentration | ORF1ab | N | RNase P |
| Human coronavirus 229E | ATCC® VR-740D | 1 x 105 copies/m L | 0/3 | 0/3 | 3/3 |
| Human coronavirus 0C43 | ATCC- VR-1558D | 1 x 105 copies/m L | 0/3 | 0/3 | 3/3 |
| Human coronavirus HKU1 | ATCC VR-3262SD | 1 x 105 copies/m L | 0/3 | 0/3 | 3/3 |
| Human coronavirus NL63 | ATCC” 3263SD | 1 x 105 copies/m L | 0/3 | 0/3 | 3/3 |
| Influenza A | VR-95DQ | 1 x 105 copies/m L | 0/3 | 0/3 | 3/3 |
| Influenza B | VR-1885DQ | 1 x 105 copies/m L | 0/3 | 0/3 | 3/3 |
| Respiratory syncytial virus | ATCC® VR-1580DQ | 1 x 105 copies/m L | 0/3 | 0/3 | 3/3 |
| Pseudomonas aeruginosa | ATCC® 27853D-5 | 1 x 106 copies/m L | 0/3 | 0/3 | 3/3 |
| Staphylococcus epidermis | ATCC’ 12228D-5 | 1 x 106 copies/m L | 0/3 | 0/3 | 3/3 |
| Candida albicans | ATCC® 10231D-5 | 1 x 106 copies/m L | 0/3 | 0/3 | 3/3 |
Endogenous Interference Substances Studies:The Sherlock ™ CRISPR SARS-CoV-2 kit uses a conventional nucleic acid extraction method; we do not anticipate interference from common endogenous substances using this method.
Clinical Evaluation:The clinical evaluation was performed on 30 contrived positive and 30 contrived negative nasopharyngeal specimens. Individual nasopharyngeal swab samples collected in the 2018 -2019 Flu season and confirmed to be negative for SARS-CoV-2 were used either unaltered (30 specimens), or spiked with extracted, quantitated SARS-CoV-2 viral genomic RNA to a concentration of 2x LoD (20 specimens), 3x LoD (5 specimens), or 5x LoD (5 specimens). For positive specimens, viral RNA was added to the nasopharyngeal matrix after the initial lysis step for the PureLink™ Viral RNA/DNA Mini Kit (Thermo Fisher) to prevent the degradation of unencapsulated RNA. The samples were randomized, then processed using the Sherlock TM CRISPR SARS-CoV-2 kit workflow. The results, as presented in Table 17 below, showed 100% agreement with the expected results for both the positive and negative specimens.
Table 17: Contrived clinical sample evaluation
| SampleConcentration | Number ofsamples | NumberDetected | % Agreement(95% confidenceinterval) |
| 5x LoD | 5 | 5 | 100%(NA*) |
| 3x LoD | 5 | 5 | 100%(NA*) |
| 2x LoD | 20 | 20 | 100%(83.9% – 100%) |
| Negativespecimens (NS) | 30 | 0 | 100%(88.6% – 100%) |
Disposal
Dispose of hazardous or biologically contaminated materials according to the practices of your institution.The product contains no substances which at their given concentration, are considered to be hazardous to health.
HMIS
| Health | 0 |
| Flammability | 0 |
| Reactivity | 0 |
Symbols Used In Packaging
| Symbol | Definition |
| Rx | Prescription Only |
| EAU | Emergency Use Authorization |
   |
In-vitro Diagnostic Use |
| Catalog Number | |
| Manufactured By | |
| Temperature Limitation | |
  |
Batch Code |
| Expiration Date | |
  |
Contains sufficient for <n> tests |
| 1011-11-17 | Date Format (year-month-day) |
| 1011-11 | Date Format (year-month) |
References
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020 Jan 24.
- World Health Organization (WHO). Coronavirus. Geneva: WHO; 2020 [Accessed 21 Jan 2020]. Available from: https://www.who.int/health-topics/coronavirus
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020 Jan 29.
- Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020 Jan 30.
- Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020 Jan 24
- Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, et al. First Case of 2019 Novel Coronavirus in the United States. N Engl J Med. 2020 Jan 31
- Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020
- Wu, F. et al. A new coronavirus associated with human respiratory disease in China.Nature, doi:10.1038/s41586-020-2008-3 (2020).
- Lu, R. et al. Genomic characterization and epidemiology of 2019 novel coronavirus:implications for virus origins and receptor binding. Lancet, doi:10.1016/S01406736(20)30251-8 (2020).
- Carmon et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR.Euro Surveill. 2020;25(3):2000045
- Lucia et al. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection 3 method based on CRISPR-Cas12. BioRxiV 2020: 1-10
CDC guidelines for Sample collection –https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html
FDA EUA guidance –https://www.fda.gov/regulatory-information/search-fda-guidance-documents/policydiagnostic-tests-coronavirus-disease-2019-during-public-health-emergency
Thermo Fisher viral RNA extraction kit PureLink™ Viral RNA/DNA Mini Kit (Cat# 12280050) Kit for RNA isolationhttps://www.thermofisher.com/order/catalog/product/12280050#/12280050 Catalog Number 12280050 Publication NumberMAN0000562
Contact Information, Ordering, and Product Support
Information and product support can be obtained from:
Contact: Sherlock Biosciences Customer SupportEmail: [email protected]Phone: 617-702-6263Website: www.sherlock.bio
Product support information
- Product FAQs
- Technical support
- Order and web support
Product documentation
- User guides, manuals, and protocols
- Fact Sheet for Healthcare Providers
- Fact Sheet for Patients
- Safety Data Sheets (SDSs; also known as MSDSs)
Note: For SDSs for reagents and chemicals from other manufacturers, contact the manufacturer.
References
[xyz-ips snippet=”download-snippet”]